Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
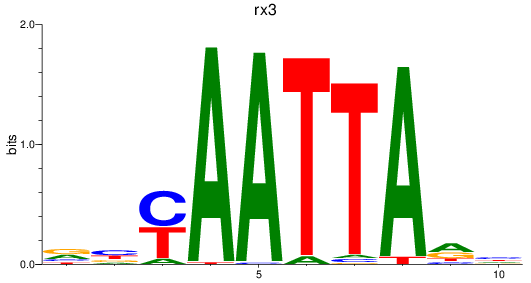
Results for rx3
Z-value: 1.45
Transcription factors associated with rx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rx3
|
ENSDARG00000052893 | retinal homeobox gene 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rx3 | dr11_v1_chr21_+_10756154_10756154 | -0.58 | 1.0e-01 | Click! |
Activity profile of rx3 motif
Sorted Z-values of rx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25777425 | 5.63 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr10_-_21362071 | 5.30 |
ENSDART00000125167
|
avd
|
avidin |
| chr10_-_21362320 | 5.21 |
ENSDART00000189789
|
avd
|
avidin |
| chr18_-_40708537 | 4.94 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr11_-_44801968 | 4.91 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr8_+_45334255 | 4.86 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr24_+_12835935 | 4.64 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr9_-_35633827 | 4.62 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr10_-_34002185 | 3.93 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr20_-_23426339 | 3.86 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr10_+_6884627 | 3.72 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr24_+_1023839 | 3.64 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr1_-_18811517 | 3.37 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr2_+_6253246 | 3.33 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr2_-_15324837 | 3.16 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr19_+_2631565 | 2.92 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr12_-_33357655 | 2.53 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr16_-_42056137 | 2.43 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr14_-_8940499 | 2.35 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr10_+_6884123 | 2.10 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr10_-_32494499 | 2.02 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr14_+_34490445 | 1.93 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr1_+_35985813 | 1.91 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr17_-_40956035 | 1.90 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr21_-_32060993 | 1.87 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr10_-_32494304 | 1.84 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr22_-_21897203 | 1.74 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr10_-_13343831 | 1.72 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr17_+_19630272 | 1.63 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr7_+_34592526 | 1.61 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr12_-_6880694 | 1.59 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr24_-_25144441 | 1.55 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr17_+_16046132 | 1.54 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr6_+_40922572 | 1.53 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr11_+_31864921 | 1.43 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr9_-_3934963 | 1.43 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr13_+_38814521 | 1.39 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr13_-_31017960 | 1.38 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr2_-_21820697 | 1.38 |
ENSDART00000135230
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr19_-_8768564 | 1.36 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr20_-_37813863 | 1.33 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr17_+_16046314 | 1.32 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr6_-_40922971 | 1.23 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr19_-_30510259 | 1.21 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr6_+_21001264 | 1.21 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr24_+_39518774 | 1.20 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr9_-_50001606 | 1.18 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr8_-_39822917 | 1.16 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr1_+_18811679 | 1.11 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr18_-_43884044 | 1.11 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr1_-_45616470 | 1.08 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr16_+_27383717 | 1.06 |
ENSDART00000132329
ENSDART00000136256 |
stx17
|
syntaxin 17 |
| chr15_+_31344472 | 1.04 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr15_-_16177603 | 1.03 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr8_+_7801060 | 1.02 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr5_-_67629263 | 1.01 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr16_+_42471455 | 1.00 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr25_-_27621268 | 0.96 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr24_+_14581864 | 0.94 |
ENSDART00000134536
|
thtpa
|
thiamine triphosphatase |
| chr3_+_13929860 | 0.93 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr15_-_43284021 | 0.89 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr6_-_12172424 | 0.89 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr23_-_17003533 | 0.88 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr8_-_19467011 | 0.88 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr20_-_40758410 | 0.87 |
ENSDART00000183031
|
cx34.5
|
connexin 34.5 |
| chr17_+_37227936 | 0.81 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr1_+_513986 | 0.81 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr9_+_50001746 | 0.81 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr20_-_45060241 | 0.80 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr5_+_66433287 | 0.79 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr11_-_29768054 | 0.74 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr7_-_23768234 | 0.72 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr21_-_15200556 | 0.69 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr10_-_2971407 | 0.68 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr14_+_8940326 | 0.67 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr5_+_32009542 | 0.66 |
ENSDART00000182025
ENSDART00000179879 |
scai
|
suppressor of cancer cell invasion |
| chr10_-_33297864 | 0.65 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr17_+_8799451 | 0.64 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr9_+_22485343 | 0.64 |
ENSDART00000146028
|
dgkg
|
diacylglycerol kinase, gamma |
| chr2_+_20793982 | 0.62 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr19_-_25119443 | 0.61 |
ENSDART00000148953
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr25_-_13490744 | 0.59 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr18_+_8320165 | 0.58 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr1_-_26444075 | 0.55 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr24_+_19415124 | 0.52 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr20_-_29864390 | 0.51 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr17_+_8799661 | 0.47 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr20_+_28803977 | 0.47 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr18_+_2228737 | 0.45 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr22_-_7129631 | 0.42 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr5_-_27438812 | 0.41 |
ENSDART00000078755
|
drd7
|
dopamine receptor D7 |
| chr17_+_10593398 | 0.36 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr17_+_12865746 | 0.35 |
ENSDART00000157083
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr14_+_40874608 | 0.32 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr14_-_7207961 | 0.31 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr12_+_48803098 | 0.29 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr23_-_1348933 | 0.28 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr14_+_23717165 | 0.28 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr17_-_200316 | 0.25 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr11_+_33312601 | 0.24 |
ENSDART00000188024
|
cntnap5l
|
contactin associated protein-like 5 like |
| chr5_+_60590796 | 0.21 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr9_+_22364997 | 0.20 |
ENSDART00000188054
ENSDART00000046116 |
crygs3
|
crystallin, gamma S3 |
| chr24_-_4782052 | 0.19 |
ENSDART00000149911
|
agtr1b
|
angiotensin II receptor, type 1b |
| chr15_-_5178899 | 0.19 |
ENSDART00000132148
|
or126-4
|
odorant receptor, family E, subfamily 126, member 4 |
| chr24_-_35282568 | 0.18 |
ENSDART00000167406
ENSDART00000088609 |
sntg1
|
syntrophin, gamma 1 |
| chr3_+_32365811 | 0.15 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr8_-_25034411 | 0.12 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr6_+_41191482 | 0.09 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr17_+_26352372 | 0.09 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr14_-_2933185 | 0.05 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr16_-_16761164 | 0.05 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr15_-_9272328 | 0.05 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr2_+_50608099 | 0.03 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr2_+_39021282 | 0.02 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr20_-_9095105 | 0.01 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
Network of associatons between targets according to the STRING database.
First level regulatory network of rx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 1.1 | 5.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.6 | 1.9 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.4 | 1.2 | GO:0097065 | anterior head development(GO:0097065) |
| 0.3 | 5.8 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 5.8 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.3 | 1.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 4.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 3.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 0.6 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 0.9 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 0.9 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.7 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 2.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 1.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 5.7 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.1 | 1.4 | GO:0048798 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 3.2 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.0 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 5.4 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.1 | 1.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 2.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.1 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 1.7 | GO:0001508 | action potential(GO:0001508) |
| 0.0 | 0.4 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.6 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.3 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.6 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 5.8 | GO:0043186 | P granule(GO:0043186) |
| 0.2 | 1.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 0.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 6.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 6.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 1.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 0.9 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 3.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.7 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 5.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 3.9 | GO:0000323 | lytic vacuole(GO:0000323) |
| 0.0 | 5.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.7 | 3.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 4.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 5.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 0.8 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 0.9 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.2 | 1.7 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 2.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.7 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 1.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 6.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.6 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 1.0 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 4.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 1.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 3.2 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 9.8 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 9.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 3.9 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.9 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.4 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |