Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
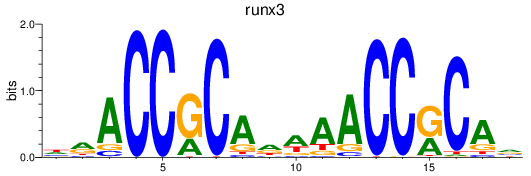
Results for runx3
Z-value: 0.41
Transcription factors associated with runx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx3
|
ENSDARG00000052826 | RUNX family transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| runx3 | dr11_v1_chr13_-_45201300_45201300 | -0.35 | 3.6e-01 | Click! |
Activity profile of runx3 motif
Sorted Z-values of runx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_50710468 | 0.70 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr6_-_9581949 | 0.58 |
ENSDART00000144335
|
cyp27c1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr16_+_28754403 | 0.46 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr15_-_20468302 | 0.43 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr2_+_4146606 | 0.42 |
ENSDART00000171170
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr8_+_17184602 | 0.42 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr16_+_23431189 | 0.41 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr19_-_5380770 | 0.41 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr6_-_2154137 | 0.39 |
ENSDART00000162656
|
tgm5l
|
transglutaminase 5, like |
| chr2_-_56348727 | 0.35 |
ENSDART00000060745
|
uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr15_-_2657508 | 0.34 |
ENSDART00000102086
|
cldna
|
claudin a |
| chr16_+_43344475 | 0.32 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr3_-_49382896 | 0.30 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr9_+_37141836 | 0.30 |
ENSDART00000024555
|
gli2a
|
GLI family zinc finger 2a |
| chr20_-_48485354 | 0.27 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr9_-_20372977 | 0.25 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr11_-_30636163 | 0.24 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr19_-_24443867 | 0.24 |
ENSDART00000163763
ENSDART00000043133 |
thbs3b
|
thrombospondin 3b |
| chr20_+_10723292 | 0.23 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr20_-_14012859 | 0.22 |
ENSDART00000152429
|
si:ch211-22i13.2
|
si:ch211-22i13.2 |
| chr2_+_53114476 | 0.22 |
ENSDART00000080850
ENSDART00000104187 |
CR384061.1
|
|
| chr5_-_10239079 | 0.21 |
ENSDART00000132739
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr14_-_24332786 | 0.21 |
ENSDART00000173164
|
fam13b
|
family with sequence similarity 13, member B |
| chr22_-_14161309 | 0.19 |
ENSDART00000133365
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr25_-_10503043 | 0.19 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr22_+_11520249 | 0.18 |
ENSDART00000063147
|
sgsh
|
N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| chr7_+_25858380 | 0.17 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr14_-_42231293 | 0.17 |
ENSDART00000185486
|
BX890543.1
|
|
| chr2_-_183992 | 0.16 |
ENSDART00000126704
ENSDART00000191283 ENSDART00000034783 |
zgc:113518
|
zgc:113518 |
| chr8_+_1065458 | 0.16 |
ENSDART00000081432
|
sprb
|
sepiapterin reductase b |
| chr16_-_7228276 | 0.16 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr24_-_14212521 | 0.15 |
ENSDART00000130825
|
xkr9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr22_+_9922301 | 0.15 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr19_-_72398 | 0.15 |
ENSDART00000165587
|
dnajc8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr20_-_53321499 | 0.15 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr18_+_16986495 | 0.15 |
ENSDART00000147377
|
si:ch211-218c6.8
|
si:ch211-218c6.8 |
| chr9_-_23747264 | 0.14 |
ENSDART00000141461
ENSDART00000010311 |
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr22_+_10757848 | 0.14 |
ENSDART00000145551
|
tmprss9
|
transmembrane protease, serine 9 |
| chr7_+_56651759 | 0.14 |
ENSDART00000073600
|
kcng4b
|
potassium voltage-gated channel, subfamily G, member 4b |
| chr22_-_30973791 | 0.14 |
ENSDART00000104728
|
ssuh2.2
|
ssu-2 homolog, tandem duplicate 2 |
| chr16_+_35870456 | 0.13 |
ENSDART00000184321
|
thrap3a
|
thyroid hormone receptor associated protein 3a |
| chr10_+_45148167 | 0.12 |
ENSDART00000172621
|
ogdhb
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) |
| chr7_-_65492048 | 0.12 |
ENSDART00000162381
|
dkk3a
|
dickkopf WNT signaling pathway inhibitor 3a |
| chr15_+_29292154 | 0.11 |
ENSDART00000137817
|
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr12_-_43711918 | 0.11 |
ENSDART00000193110
|
CU914622.2
|
|
| chr14_-_16476863 | 0.10 |
ENSDART00000089021
|
canx
|
calnexin |
| chr17_-_27382826 | 0.10 |
ENSDART00000186657
ENSDART00000155986 ENSDART00000191060 ENSDART00000077608 |
si:ch1073-358c10.1
|
si:ch1073-358c10.1 |
| chr1_+_54911458 | 0.10 |
ENSDART00000089603
|
golga7ba
|
golgin A7 family, member Ba |
| chr24_-_14711597 | 0.10 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr19_+_42219165 | 0.09 |
ENSDART00000163192
|
CU896644.1
|
|
| chr13_-_17729474 | 0.09 |
ENSDART00000013011
|
vdac2
|
voltage-dependent anion channel 2 |
| chr8_-_17184482 | 0.08 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr11_-_17755444 | 0.08 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr1_-_43727418 | 0.08 |
ENSDART00000133715
ENSDART00000074597 ENSDART00000132542 ENSDART00000181792 |
bdh2
SLC9B2
|
3-hydroxybutyrate dehydrogenase, type 2 si:dkey-162b23.4 |
| chr18_-_50524017 | 0.08 |
ENSDART00000150013
ENSDART00000149912 |
cd276
|
CD276 molecule |
| chr24_+_24064562 | 0.08 |
ENSDART00000144394
|
zgc:112408
|
zgc:112408 |
| chr23_-_36313431 | 0.08 |
ENSDART00000125860
|
nfe2
|
nuclear factor, erythroid 2 |
| chr10_+_45148005 | 0.07 |
ENSDART00000182501
|
ogdhb
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) |
| chr3_+_14339728 | 0.07 |
ENSDART00000184127
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr18_+_808911 | 0.07 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr3_-_22228602 | 0.07 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr23_+_20795781 | 0.07 |
ENSDART00000128577
|
ttc34
|
tetratricopeptide repeat domain 34 |
| chr5_-_71995108 | 0.07 |
ENSDART00000124587
|
fam78ab
|
family with sequence similarity 78, member Ab |
| chr25_-_225964 | 0.07 |
ENSDART00000193424
|
CABZ01113818.1
|
|
| chr15_-_31067589 | 0.06 |
ENSDART00000060157
|
lgals9l3
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 3 |
| chr12_-_43819253 | 0.06 |
ENSDART00000160261
ENSDART00000170045 ENSDART00000159106 |
si:ch73-329n5.6
|
si:ch73-329n5.6 |
| chr23_+_36308717 | 0.06 |
ENSDART00000131711
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr17_+_7522777 | 0.06 |
ENSDART00000184723
|
klhl10b.2
|
kelch-like family member 10b, tandem duplicate 2 |
| chr20_-_2725930 | 0.06 |
ENSDART00000081643
|
akirin2
|
akirin 2 |
| chr21_-_43992027 | 0.06 |
ENSDART00000188612
|
cdx1b
|
caudal type homeobox 1 b |
| chr18_-_50523399 | 0.05 |
ENSDART00000033591
|
cd276
|
CD276 molecule |
| chr15_+_37589698 | 0.05 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr19_+_2590182 | 0.05 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr1_+_16396870 | 0.05 |
ENSDART00000189245
ENSDART00000113266 |
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr10_+_21656654 | 0.05 |
ENSDART00000160464
|
pcdh1g2
|
protocadherin 1 gamma 2 |
| chr22_-_25502977 | 0.05 |
ENSDART00000181749
|
CR769772.4
|
|
| chr22_+_11144153 | 0.05 |
ENSDART00000047442
|
bcor
|
BCL6 corepressor |
| chr1_+_16397063 | 0.04 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr12_+_46883785 | 0.04 |
ENSDART00000008312
|
fam53b
|
family with sequence similarity 53, member B |
| chr11_-_8208464 | 0.04 |
ENSDART00000161283
|
pimr203
|
Pim proto-oncogene, serine/threonine kinase, related 203 |
| chr21_+_15824182 | 0.04 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr1_+_39859782 | 0.04 |
ENSDART00000149984
|
irf2a
|
interferon regulatory factor 2a |
| chr6_-_11768198 | 0.04 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr11_-_40519886 | 0.04 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr25_-_210730 | 0.03 |
ENSDART00000187580
|
FP236318.1
|
|
| chr22_-_25469751 | 0.03 |
ENSDART00000171670
|
CR769772.4
|
|
| chr13_+_8255106 | 0.03 |
ENSDART00000080465
|
hells
|
helicase, lymphoid specific |
| chr6_+_25282671 | 0.03 |
ENSDART00000157575
|
si:ch211-180m24.3
|
si:ch211-180m24.3 |
| chr2_-_51521079 | 0.03 |
ENSDART00000165504
|
LO018493.1
|
|
| chr2_+_49864219 | 0.03 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr12_-_23009312 | 0.03 |
ENSDART00000111801
|
mkxa
|
mohawk homeobox a |
| chr10_-_309894 | 0.03 |
ENSDART00000163287
|
CABZ01049607.1
|
|
| chr9_+_34148714 | 0.03 |
ENSDART00000078051
|
gpr161
|
G protein-coupled receptor 161 |
| chr6_-_141564 | 0.03 |
ENSDART00000151245
ENSDART00000063876 |
s1pr5b
|
sphingosine-1-phosphate receptor 5b |
| chr22_+_17203752 | 0.03 |
ENSDART00000143376
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr5_+_7989210 | 0.02 |
ENSDART00000168071
|
gdnfb
|
glial cell derived neurotrophic factor b |
| chr1_-_8012476 | 0.02 |
ENSDART00000177051
|
CR855320.3
|
|
| chr15_-_4037626 | 0.01 |
ENSDART00000160905
|
P2RY14
|
purinergic receptor P2Y14 |
| chr3_+_6291635 | 0.01 |
ENSDART00000185055
ENSDART00000157707 |
si:ch211-12p12.2
|
si:ch211-12p12.2 |
| chr1_+_23408622 | 0.00 |
ENSDART00000140706
|
chrna9
|
cholinergic receptor, nicotinic, alpha 9 |
| chr19_-_40906161 | 0.00 |
ENSDART00000004514
|
bet1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr6_+_44163727 | 0.00 |
ENSDART00000064878
|
gxylt2
|
glucoside xylosyltransferase 2 |
| chr12_+_28910762 | 0.00 |
ENSDART00000076342
ENSDART00000160939 ENSDART00000076572 |
rnf40
|
ring finger protein 40 |
| chr22_-_392224 | 0.00 |
ENSDART00000124720
|
CABZ01072989.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of runx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.1 | 0.3 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0055057 | neuroblast division(GO:0055057) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.1 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.0 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |