Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
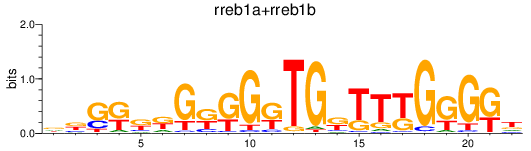
Results for rreb1a+rreb1b
Z-value: 1.19
Transcription factors associated with rreb1a+rreb1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rreb1b
|
ENSDARG00000042652 | ras responsive element binding protein 1b |
|
rreb1a
|
ENSDARG00000063701 | ras responsive element binding protein 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rreb1a | dr11_v1_chr24_-_2423791_2423791 | -0.65 | 5.6e-02 | Click! |
| rreb1b | dr11_v1_chr2_+_21048661_21048661 | -0.57 | 1.1e-01 | Click! |
Activity profile of rreb1a+rreb1b motif
Sorted Z-values of rreb1a+rreb1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_56351 | 2.76 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr2_-_22535 | 2.53 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr7_-_2039060 | 2.40 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr22_-_34872533 | 2.40 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr15_+_47161917 | 1.31 |
ENSDART00000167860
|
gap43
|
growth associated protein 43 |
| chr19_-_5358443 | 1.30 |
ENSDART00000105036
|
cyt1l
|
type I cytokeratin, enveloping layer, like |
| chr19_-_5364649 | 1.27 |
ENSDART00000004812
|
cyt1
|
type I cytokeratin, enveloping layer |
| chr20_-_54462551 | 1.26 |
ENSDART00000171769
ENSDART00000169692 |
evlb
|
Enah/Vasp-like b |
| chr8_+_54284961 | 1.24 |
ENSDART00000122692
|
plxnd1
|
plexin D1 |
| chr17_-_122680 | 1.23 |
ENSDART00000066430
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr12_+_42436328 | 1.22 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr21_+_28445052 | 1.18 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr22_+_10090673 | 1.13 |
ENSDART00000186680
|
si:dkey-102c8.3
|
si:dkey-102c8.3 |
| chr25_-_173165 | 1.11 |
ENSDART00000193594
|
CABZ01114053.1
|
|
| chr4_-_16345227 | 1.02 |
ENSDART00000079521
|
kera
|
keratocan |
| chr9_+_30294096 | 0.99 |
ENSDART00000026551
|
srpx
|
sushi-repeat containing protein, X-linked |
| chr3_-_5664123 | 0.98 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr14_-_9522364 | 0.97 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr19_+_42983613 | 0.95 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr7_-_73752955 | 0.93 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr19_+_30633453 | 0.93 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr13_-_638485 | 0.90 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr7_-_74090168 | 0.88 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr5_-_72125551 | 0.87 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr9_+_21722733 | 0.86 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr2_-_44183613 | 0.83 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr4_+_77735212 | 0.80 |
ENSDART00000160716
|
si:dkey-238k10.1
|
si:dkey-238k10.1 |
| chr2_-_44183451 | 0.79 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr21_+_19834072 | 0.77 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr23_+_44745317 | 0.75 |
ENSDART00000165654
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr2_-_51700709 | 0.74 |
ENSDART00000188601
|
tgm1l1
|
transglutaminase 1 like 1 |
| chr13_-_11536951 | 0.74 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr13_-_31435137 | 0.74 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr8_+_6954984 | 0.74 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr10_-_35542071 | 0.73 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr21_-_41305748 | 0.73 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr1_-_9104631 | 0.73 |
ENSDART00000146642
|
si:ch211-14k19.8
|
si:ch211-14k19.8 |
| chr1_+_59538755 | 0.73 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr23_-_9768700 | 0.72 |
ENSDART00000045126
|
lama5
|
laminin, alpha 5 |
| chr20_-_10120442 | 0.72 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr9_-_23033818 | 0.72 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr2_+_33368414 | 0.72 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr15_-_452347 | 0.72 |
ENSDART00000115233
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr12_+_13244149 | 0.72 |
ENSDART00000186984
ENSDART00000105896 |
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr5_-_5147220 | 0.69 |
ENSDART00000187026
ENSDART00000162334 |
lmx1ba
|
LIM homeobox transcription factor 1, beta a |
| chr17_+_53311618 | 0.68 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr19_-_5380770 | 0.68 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr25_+_7982979 | 0.67 |
ENSDART00000171904
|
ucmab
|
upper zone of growth plate and cartilage matrix associated b |
| chr6_-_60031693 | 0.67 |
ENSDART00000160275
|
CABZ01079262.1
|
|
| chr6_-_18976168 | 0.66 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr23_+_44732863 | 0.66 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr6_+_59818275 | 0.64 |
ENSDART00000165213
|
caskb
|
calcium/calmodulin-dependent serine protein kinase b |
| chr19_-_45960191 | 0.62 |
ENSDART00000052434
ENSDART00000172732 |
eif3hb
|
eukaryotic translation initiation factor 3, subunit H, b |
| chr6_+_59808677 | 0.62 |
ENSDART00000164469
|
caskb
|
calcium/calmodulin-dependent serine protein kinase b |
| chr3_-_38692920 | 0.61 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr15_-_42736433 | 0.60 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr23_-_15284757 | 0.59 |
ENSDART00000139135
|
sulf2b
|
sulfatase 2b |
| chr4_+_76619791 | 0.59 |
ENSDART00000184042
|
ms4a17a.8
|
membrane-spanning 4-domains, subfamily A, member 17A.8 |
| chr17_-_20711735 | 0.58 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr14_+_34514336 | 0.57 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr16_+_22587661 | 0.57 |
ENSDART00000129612
ENSDART00000142241 |
she
|
Src homology 2 domain containing E |
| chr12_+_2677303 | 0.57 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr25_+_37480285 | 0.56 |
ENSDART00000166187
|
CABZ01095001.1
|
|
| chr16_-_54405976 | 0.54 |
ENSDART00000055395
|
osr2
|
odd-skipped related transciption factor 2 |
| chr13_+_22675802 | 0.54 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr18_-_8889382 | 0.53 |
ENSDART00000058647
ENSDART00000184944 |
si:dkey-95h12.1
|
si:dkey-95h12.1 |
| chr11_-_97817 | 0.53 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr23_+_3591690 | 0.51 |
ENSDART00000180822
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr1_+_59533317 | 0.51 |
ENSDART00000166821
|
sp6
|
Sp6 transcription factor |
| chr17_+_52822422 | 0.51 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr5_+_23630384 | 0.51 |
ENSDART00000013745
|
cx39.9
|
connexin 39.9 |
| chr3_+_23703704 | 0.51 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr6_-_53426773 | 0.51 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr13_-_45523026 | 0.51 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr2_-_50298337 | 0.50 |
ENSDART00000155125
|
cntnap2b
|
contactin associated protein like 2b |
| chr8_+_37489495 | 0.50 |
ENSDART00000141516
|
fmodb
|
fibromodulin b |
| chr7_+_39634873 | 0.50 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr7_-_57949117 | 0.50 |
ENSDART00000138803
|
ank2b
|
ankyrin 2b, neuronal |
| chr3_-_1190132 | 0.49 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr22_+_661505 | 0.49 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr21_-_45613564 | 0.48 |
ENSDART00000160324
|
LO018363.1
|
|
| chr6_+_59967994 | 0.48 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr1_-_59251874 | 0.48 |
ENSDART00000168919
|
olfm2b
|
olfactomedin 2b |
| chr10_+_16756546 | 0.47 |
ENSDART00000168624
|
CABZ01024659.1
|
|
| chr6_-_21189295 | 0.47 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr8_+_4368534 | 0.46 |
ENSDART00000015214
|
pptc7a
|
PTC7 protein phosphatase homolog a |
| chr4_-_193762 | 0.46 |
ENSDART00000169667
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr17_+_15388479 | 0.46 |
ENSDART00000052439
|
si:ch211-266g18.6
|
si:ch211-266g18.6 |
| chr25_+_1732838 | 0.46 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr23_-_4855122 | 0.45 |
ENSDART00000133701
|
slc6a1a
|
solute carrier family 6 (neurotransmitter transporter), member 1a |
| chr9_+_36946340 | 0.45 |
ENSDART00000135281
|
si:dkey-3d4.3
|
si:dkey-3d4.3 |
| chr19_+_233143 | 0.45 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr7_+_31838320 | 0.45 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr1_+_47585700 | 0.44 |
ENSDART00000153746
ENSDART00000084457 |
sh3pxd2aa
|
SH3 and PX domains 2Aa |
| chr16_+_10918252 | 0.44 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr11_+_38280454 | 0.43 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr13_+_41022502 | 0.43 |
ENSDART00000026808
|
dkk1a
|
dickkopf WNT signaling pathway inhibitor 1a |
| chr13_+_255067 | 0.43 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr15_-_40175894 | 0.43 |
ENSDART00000156632
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr25_+_5044780 | 0.43 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr10_+_22772084 | 0.42 |
ENSDART00000144845
ENSDART00000165222 |
tmem88a
|
transmembrane protein 88 a |
| chr25_+_258883 | 0.42 |
ENSDART00000155256
|
zgc:92481
|
zgc:92481 |
| chr25_+_25464630 | 0.41 |
ENSDART00000150537
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr10_-_24343507 | 0.41 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr5_-_28029558 | 0.41 |
ENSDART00000078649
|
abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr3_+_60007703 | 0.41 |
ENSDART00000157351
ENSDART00000153928 ENSDART00000155876 |
si:ch211-110e21.3
|
si:ch211-110e21.3 |
| chr11_-_3334248 | 0.41 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr8_-_1909840 | 0.41 |
ENSDART00000147408
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr25_-_37501371 | 0.40 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr14_+_52563794 | 0.40 |
ENSDART00000168874
|
rpl26
|
ribosomal protein L26 |
| chr1_-_5543065 | 0.39 |
ENSDART00000103755
|
fn1b
|
fibronectin 1b |
| chr17_+_51517750 | 0.39 |
ENSDART00000180896
ENSDART00000193528 |
pxdn
|
peroxidasin |
| chr18_+_45990394 | 0.38 |
ENSDART00000024068
|
mmp23bb
|
matrix metallopeptidase 23bb |
| chr20_-_35246150 | 0.38 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr2_+_28889936 | 0.37 |
ENSDART00000078232
|
cdh10a
|
cadherin 10, type 2a (T2-cadherin) |
| chr1_-_625875 | 0.37 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr4_+_5798223 | 0.37 |
ENSDART00000059440
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr2_+_22694382 | 0.37 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr25_-_10564721 | 0.37 |
ENSDART00000154776
|
galn
|
galanin/GMAP prepropeptide |
| chr23_+_3587230 | 0.36 |
ENSDART00000055103
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr10_-_43294933 | 0.36 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr3_-_30685401 | 0.36 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr20_+_52595220 | 0.36 |
ENSDART00000180610
|
si:dkey-235d18.5
|
si:dkey-235d18.5 |
| chr20_+_4392687 | 0.36 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr18_+_21122818 | 0.35 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr18_-_81280 | 0.35 |
ENSDART00000149810
ENSDART00000149321 |
hdc
|
histidine decarboxylase |
| chr10_+_3875716 | 0.35 |
ENSDART00000189268
ENSDART00000180624 |
TTC28
|
tetratricopeptide repeat domain 28 |
| chr21_+_25765734 | 0.35 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr15_-_12500938 | 0.34 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr23_+_44580254 | 0.34 |
ENSDART00000185069
|
pfn1
|
profilin 1 |
| chr10_+_8527196 | 0.34 |
ENSDART00000141147
|
si:ch211-193e13.5
|
si:ch211-193e13.5 |
| chr16_+_68069 | 0.34 |
ENSDART00000185385
ENSDART00000159652 |
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr21_+_53504 | 0.34 |
ENSDART00000170452
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr12_+_1398404 | 0.33 |
ENSDART00000026303
|
rasd1
|
RAS, dexamethasone-induced 1 |
| chr12_+_2676878 | 0.33 |
ENSDART00000185909
|
antxr1c
|
anthrax toxin receptor 1c |
| chr23_+_3616224 | 0.33 |
ENSDART00000190917
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr7_+_35229645 | 0.33 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr7_+_36552725 | 0.32 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr19_-_47456787 | 0.32 |
ENSDART00000168792
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr25_+_5288665 | 0.32 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr5_+_71802014 | 0.32 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr14_-_853176 | 0.32 |
ENSDART00000041988
|
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr22_+_1028724 | 0.31 |
ENSDART00000149625
|
si:ch73-352p18.4
|
si:ch73-352p18.4 |
| chr24_-_40860603 | 0.31 |
ENSDART00000188032
|
CU633479.7
|
|
| chr16_-_54640315 | 0.31 |
ENSDART00000101207
|
spag1b
|
sperm associated antigen 1b |
| chr12_-_1951233 | 0.31 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr5_-_5147041 | 0.30 |
ENSDART00000180236
|
lmx1ba
|
LIM homeobox transcription factor 1, beta a |
| chr6_-_48311 | 0.30 |
ENSDART00000131010
|
zgc:114175
|
zgc:114175 |
| chr18_-_49078428 | 0.30 |
ENSDART00000160702
ENSDART00000174103 |
BX663503.3
|
|
| chr1_+_2129164 | 0.30 |
ENSDART00000074923
ENSDART00000124534 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr23_-_969844 | 0.30 |
ENSDART00000127037
|
cdh26.2
|
cadherin 26, tandem duplicate 2 |
| chr8_+_68864 | 0.30 |
ENSDART00000164574
|
prr16
|
proline rich 16 |
| chr16_+_41826584 | 0.30 |
ENSDART00000147523
|
si:dkey-199f5.6
|
si:dkey-199f5.6 |
| chr25_-_10565006 | 0.30 |
ENSDART00000130608
ENSDART00000190212 |
galn
|
galanin/GMAP prepropeptide |
| chr18_+_1154189 | 0.29 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr5_-_71838520 | 0.29 |
ENSDART00000174396
|
CU927890.1
|
|
| chr1_-_8012476 | 0.29 |
ENSDART00000177051
|
CR855320.3
|
|
| chr12_-_4540564 | 0.29 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr19_+_32856907 | 0.29 |
ENSDART00000148232
|
rpl30
|
ribosomal protein L30 |
| chr19_+_2279051 | 0.29 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr25_-_37489917 | 0.28 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr7_+_65876335 | 0.28 |
ENSDART00000150143
|
tead1b
|
TEA domain family member 1b |
| chr6_-_346084 | 0.28 |
ENSDART00000162599
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr21_-_44009169 | 0.28 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr17_-_52643970 | 0.27 |
ENSDART00000190594
|
spred1
|
sprouty-related, EVH1 domain containing 1 |
| chr24_-_31439841 | 0.27 |
ENSDART00000169952
|
cngb3.2
|
cyclic nucleotide gated channel beta 3, tandem duplicate 2 |
| chr16_+_43347966 | 0.27 |
ENSDART00000171308
|
zmp:0000000930
|
zmp:0000000930 |
| chr5_-_201600 | 0.27 |
ENSDART00000158495
|
CABZ01088906.1
|
|
| chr23_+_44497701 | 0.26 |
ENSDART00000149903
|
si:ch73-375g18.1
|
si:ch73-375g18.1 |
| chr18_+_22793743 | 0.26 |
ENSDART00000150106
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr13_+_51853716 | 0.26 |
ENSDART00000175341
ENSDART00000187855 |
LT631684.1
|
|
| chr19_-_13923091 | 0.26 |
ENSDART00000169993
ENSDART00000158992 |
stx12
|
syntaxin 12 |
| chr3_+_18398876 | 0.26 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr5_+_480119 | 0.25 |
ENSDART00000055681
|
tek
|
TEK tyrosine kinase, endothelial |
| chr3_-_62380146 | 0.25 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr21_+_25660613 | 0.25 |
ENSDART00000134017
|
si:dkey-17e16.15
|
si:dkey-17e16.15 |
| chr22_+_1186866 | 0.25 |
ENSDART00000167710
|
eps8l3a
|
EPS8-like 3a |
| chr23_-_38054 | 0.25 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr21_+_6151953 | 0.25 |
ENSDART00000131854
|
si:dkey-93m18.6
|
si:dkey-93m18.6 |
| chr5_-_26181863 | 0.25 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr5_-_16351306 | 0.24 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr5_-_43959972 | 0.24 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr11_-_4235811 | 0.24 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr1_+_109798 | 0.24 |
ENSDART00000165402
|
F7 (1 of many)
|
zgc:163025 |
| chr15_-_5742531 | 0.23 |
ENSDART00000045985
|
phkg1a
|
phosphorylase kinase, gamma 1a (muscle) |
| chr7_-_5191797 | 0.23 |
ENSDART00000172955
|
si:ch73-223f5.1
|
si:ch73-223f5.1 |
| chr4_+_20263097 | 0.23 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr18_-_45617146 | 0.23 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr6_+_38381957 | 0.23 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr7_+_50849142 | 0.23 |
ENSDART00000073806
|
pcolceb
|
procollagen C-endopeptidase enhancer b |
| chr4_-_6459863 | 0.22 |
ENSDART00000138367
|
foxp2
|
forkhead box P2 |
| chr12_+_10053852 | 0.22 |
ENSDART00000078497
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr19_+_43256986 | 0.22 |
ENSDART00000182336
|
DGKD
|
diacylglycerol kinase delta |
| chr25_+_37268900 | 0.22 |
ENSDART00000156737
|
si:dkey-234i14.6
|
si:dkey-234i14.6 |
| chr12_-_48992527 | 0.22 |
ENSDART00000169696
|
CDHR1 (1 of many)
|
cadherin related family member 1 |
| chr7_+_72630369 | 0.22 |
ENSDART00000170698
|
FO905040.1
|
|
| chr5_+_20589472 | 0.22 |
ENSDART00000130911
|
cmklr1
|
chemokine-like receptor 1 |
| chr13_-_20519001 | 0.22 |
ENSDART00000168955
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr17_+_8925232 | 0.22 |
ENSDART00000036668
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr4_+_76722754 | 0.21 |
ENSDART00000153867
|
ms4a17a.3
|
membrane-spanning 4-domains, subfamily A, member 17A.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rreb1a+rreb1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.3 | 1.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 0.9 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 0.7 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.6 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.4 | GO:0060923 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.4 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 1.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.5 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 0.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.9 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 1.2 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 2.8 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.1 | 1.0 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 1.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 1.4 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 1.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.7 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.3 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 1.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0043703 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.4 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.4 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.7 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.1 | GO:0035739 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 1.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:0009750 | response to fructose(GO:0009750) |
| 0.0 | 0.1 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.0 | 0.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0090183 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.3 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.6 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.4 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0006837 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.2 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.1 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.4 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.5 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.2 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 1.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.9 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 3.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.6 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.7 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.4 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.1 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.3 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 1.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.4 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 0.4 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.6 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.6 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |