Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
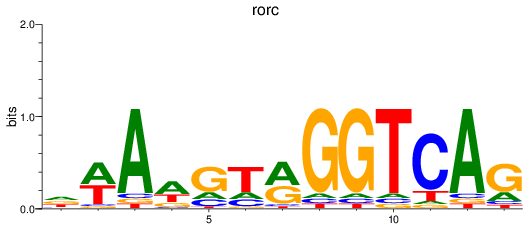
Results for rorc
Z-value: 0.54
Transcription factors associated with rorc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rorc
|
ENSDARG00000087195 | RAR-related orphan receptor C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rorc | dr11_v1_chr16_+_17913936_17913945 | 0.08 | 8.4e-01 | Click! |
Activity profile of rorc motif
Sorted Z-values of rorc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_43314700 | 0.44 |
ENSDART00000049045
ENSDART00000133158 |
matn1
|
matrilin 1 |
| chr5_+_70155935 | 0.23 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr8_+_30452945 | 0.23 |
ENSDART00000062303
|
foxd5
|
forkhead box D5 |
| chr16_+_26738404 | 0.22 |
ENSDART00000163110
|
fsbp
|
fibrinogen silencer binding protein |
| chr15_+_19682013 | 0.22 |
ENSDART00000127368
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr18_+_7594012 | 0.19 |
ENSDART00000062150
|
zgc:77752
|
zgc:77752 |
| chr15_+_19681718 | 0.18 |
ENSDART00000164803
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr7_+_48675347 | 0.17 |
ENSDART00000157917
ENSDART00000185580 |
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr19_+_42336523 | 0.13 |
ENSDART00000151304
|
anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr15_-_43625549 | 0.13 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr15_+_42599501 | 0.13 |
ENSDART00000177646
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr11_-_1948784 | 0.13 |
ENSDART00000082475
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr10_+_2232023 | 0.12 |
ENSDART00000097695
|
cntnap3
|
contactin associated protein like 3 |
| chr15_-_37829160 | 0.12 |
ENSDART00000099425
|
ctrl
|
chymotrypsin-like |
| chr10_+_26935155 | 0.12 |
ENSDART00000129993
|
CR450793.1
|
|
| chr17_-_13072334 | 0.11 |
ENSDART00000159598
|
CU469462.1
|
|
| chr1_+_19433004 | 0.11 |
ENSDART00000133959
|
clockb
|
clock circadian regulator b |
| chr7_-_22632518 | 0.11 |
ENSDART00000161046
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr19_+_30990815 | 0.10 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr2_+_27867830 | 0.10 |
ENSDART00000128193
|
rnf152
|
ring finger protein 152 |
| chr22_-_13544244 | 0.10 |
ENSDART00000110136
|
cntnap5b
|
contactin associated protein-like 5b |
| chr3_-_26191960 | 0.10 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr13_+_29470442 | 0.09 |
ENSDART00000028417
|
lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr19_+_24896409 | 0.09 |
ENSDART00000049840
|
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr7_-_22632938 | 0.09 |
ENSDART00000159867
ENSDART00000165706 |
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr21_+_27189490 | 0.09 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr5_+_2815021 | 0.09 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr17_+_2214283 | 0.09 |
ENSDART00000189187
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr2_+_28889936 | 0.09 |
ENSDART00000078232
|
cdh10a
|
cadherin 10, type 2a (T2-cadherin) |
| chr12_-_7234915 | 0.09 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr12_+_31616412 | 0.08 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr4_+_17280868 | 0.08 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr18_-_42313798 | 0.08 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr11_-_24458786 | 0.08 |
ENSDART00000089713
|
mxra8a
|
matrix-remodelling associated 8a |
| chr11_-_42396302 | 0.08 |
ENSDART00000165624
|
slmapa
|
sarcolemma associated protein a |
| chr8_+_42917515 | 0.08 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr23_+_44741500 | 0.08 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr7_+_24573721 | 0.08 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr5_+_19413838 | 0.07 |
ENSDART00000089014
ENSDART00000146215 |
myo1ha
|
myosin IHa |
| chr14_+_17376940 | 0.07 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr3_+_60761811 | 0.07 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr5_-_16983336 | 0.07 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr22_-_18546241 | 0.07 |
ENSDART00000105404
ENSDART00000105405 |
cirbpb
|
cold inducible RNA binding protein b |
| chr9_+_13682133 | 0.07 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr19_+_30990129 | 0.07 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr16_+_16266428 | 0.07 |
ENSDART00000188433
|
setd2
|
SET domain containing 2 |
| chr14_-_42997145 | 0.07 |
ENSDART00000172801
|
pcdh10b
|
protocadherin 10b |
| chr19_-_8604429 | 0.06 |
ENSDART00000151165
|
trim46b
|
tripartite motif containing 46b |
| chr17_-_37312922 | 0.06 |
ENSDART00000153555
|
si:ch211-180n24.3
|
si:ch211-180n24.3 |
| chr5_-_11573490 | 0.06 |
ENSDART00000109577
|
FO704871.1
|
|
| chr8_+_25351863 | 0.06 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr10_+_575929 | 0.06 |
ENSDART00000129856
|
smad4a
|
SMAD family member 4a |
| chr15_-_31419805 | 0.06 |
ENSDART00000060111
|
or111-11
|
odorant receptor, family D, subfamily 111, member 11 |
| chr10_+_15454745 | 0.05 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr3_-_5555500 | 0.05 |
ENSDART00000176133
ENSDART00000057464 |
trim35-31
|
tripartite motif containing 35-31 |
| chr7_+_65240227 | 0.05 |
ENSDART00000168287
|
bco1l
|
beta-carotene oxygenase 1, like |
| chr2_+_32743807 | 0.05 |
ENSDART00000022909
|
klhl18
|
kelch-like family member 18 |
| chr6_+_13742899 | 0.05 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr23_+_19564392 | 0.05 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr7_+_19835569 | 0.05 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr2_+_49417900 | 0.05 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr2_-_38337122 | 0.05 |
ENSDART00000076523
ENSDART00000187473 |
slc7a8b
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8b |
| chr8_+_25352268 | 0.05 |
ENSDART00000187829
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr25_-_19219807 | 0.05 |
ENSDART00000183577
|
acanb
|
aggrecan b |
| chr2_-_56649883 | 0.05 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr23_-_36724575 | 0.05 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr7_+_22313533 | 0.05 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr16_+_12632428 | 0.05 |
ENSDART00000184600
ENSDART00000180537 |
nat14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr7_-_28058442 | 0.05 |
ENSDART00000173842
|
si:ch211-235p24.2
|
si:ch211-235p24.2 |
| chr19_-_8096984 | 0.05 |
ENSDART00000146987
|
si:dkey-266f7.9
|
si:dkey-266f7.9 |
| chr8_+_15277874 | 0.05 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr10_+_44692272 | 0.05 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr15_-_34322915 | 0.05 |
ENSDART00000190543
ENSDART00000019651 ENSDART00000193601 |
dgkb
|
diacylglycerol kinase, beta |
| chr8_+_3649036 | 0.05 |
ENSDART00000033152
|
rab35b
|
RAB35, member RAS oncogene family b |
| chr12_-_32013125 | 0.05 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr11_+_30672282 | 0.04 |
ENSDART00000023981
ENSDART00000188846 |
ttbk1a
|
tau tubulin kinase 1a |
| chr8_-_51349429 | 0.04 |
ENSDART00000134043
ENSDART00000098282 |
si:ch211-198o12.4
|
si:ch211-198o12.4 |
| chr9_-_43207768 | 0.04 |
ENSDART00000192523
|
sestd1
|
SEC14 and spectrin domains 1 |
| chr11_+_26363435 | 0.04 |
ENSDART00000088800
|
ppcs
|
phosphopantothenoylcysteine synthetase |
| chr14_-_25111496 | 0.04 |
ENSDART00000158108
|
si:rp71-1d10.8
|
si:rp71-1d10.8 |
| chr25_+_3035384 | 0.04 |
ENSDART00000184219
ENSDART00000149360 |
mpi
|
mannose phosphate isomerase |
| chr1_+_9708801 | 0.04 |
ENSDART00000189621
|
elfn1b
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1b |
| chr15_+_1766734 | 0.04 |
ENSDART00000168250
|
cul3b
|
cullin 3b |
| chr8_+_18463797 | 0.04 |
ENSDART00000148958
|
alkal1
|
ALK and LTK ligand 1 |
| chr8_+_14890821 | 0.03 |
ENSDART00000190966
|
soat1
|
sterol O-acyltransferase 1 |
| chr25_-_37227491 | 0.03 |
ENSDART00000156647
|
glg1a
|
golgi glycoprotein 1a |
| chr3_+_13637383 | 0.03 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr8_+_26034623 | 0.03 |
ENSDART00000004521
ENSDART00000142555 |
arih2
|
ariadne homolog 2 (Drosophila) |
| chr22_-_621609 | 0.03 |
ENSDART00000137264
ENSDART00000106636 |
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr8_-_29706882 | 0.03 |
ENSDART00000045909
|
prf1.5
|
perforin 1.5 |
| chr16_+_11242443 | 0.03 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr24_+_24809877 | 0.03 |
ENSDART00000185117
|
dnajc5b
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr15_-_25304710 | 0.03 |
ENSDART00000190690
|
pafah1b1a
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1a |
| chr3_-_31226841 | 0.03 |
ENSDART00000085903
|
syt17
|
synaptotagmin XVII |
| chr15_-_34878388 | 0.02 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr22_-_38480186 | 0.02 |
ENSDART00000171704
|
soul4
|
heme-binding protein soul4 |
| chr17_-_6738538 | 0.02 |
ENSDART00000157125
|
vsnl1b
|
visinin-like 1b |
| chr4_+_38103421 | 0.02 |
ENSDART00000164164
|
znf1016
|
zinc finger protein 1016 |
| chr16_+_16265850 | 0.02 |
ENSDART00000181265
|
setd2
|
SET domain containing 2 |
| chr9_+_21993092 | 0.02 |
ENSDART00000059693
|
crygm7
|
crystallin, gamma M7 |
| chr15_+_17100697 | 0.02 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr20_-_14680897 | 0.02 |
ENSDART00000063857
ENSDART00000161314 |
scrn2
|
secernin 2 |
| chr22_-_6420239 | 0.02 |
ENSDART00000148385
|
zgc:171699
|
zgc:171699 |
| chr2_+_49864219 | 0.02 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr5_-_69523816 | 0.02 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
| chr5_-_30079434 | 0.02 |
ENSDART00000133981
|
bco2a
|
beta-carotene oxygenase 2a |
| chr8_-_25771474 | 0.02 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr9_+_35876927 | 0.02 |
ENSDART00000138834
|
mab21l3
|
mab-21-like 3 |
| chr1_+_17527931 | 0.02 |
ENSDART00000131927
|
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr10_+_40548616 | 0.02 |
ENSDART00000147894
|
si:ch211-238p8.10
|
si:ch211-238p8.10 |
| chr1_-_21714025 | 0.02 |
ENSDART00000129066
|
zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr1_-_33353778 | 0.02 |
ENSDART00000041191
|
gyg2
|
glycogenin 2 |
| chr11_+_19159083 | 0.02 |
ENSDART00000138964
|
adamts9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr11_-_39118882 | 0.02 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr22_+_26263290 | 0.02 |
ENSDART00000184840
|
BX649315.1
|
|
| chr15_-_2841677 | 0.02 |
ENSDART00000026145
ENSDART00000180290 |
AMOTL1
|
angiomotin like 1 |
| chr14_-_22390821 | 0.02 |
ENSDART00000054396
|
fbxl3l
|
F-box and leucine-rich repeat protein 3, like |
| chr17_-_2126517 | 0.02 |
ENSDART00000013093
|
zfyve19
|
zinc finger, FYVE domain containing 19 |
| chr25_+_11389220 | 0.01 |
ENSDART00000187787
|
AGBL1
|
si:ch73-141f14.1 |
| chr24_-_35707552 | 0.01 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr12_+_30789611 | 0.01 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr13_-_1130096 | 0.01 |
ENSDART00000010261
|
pno1
|
partner of NOB1 homolog |
| chr22_+_8612462 | 0.01 |
ENSDART00000114586
|
CR450686.1
|
|
| chr6_-_8244474 | 0.01 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr1_+_15204633 | 0.01 |
ENSDART00000188410
|
itln1
|
intelectin 1 |
| chr22_-_834106 | 0.01 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr8_+_43340995 | 0.01 |
ENSDART00000038566
|
rflna
|
refilin A |
| chr17_-_31044803 | 0.01 |
ENSDART00000185941
ENSDART00000152016 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr16_+_30438041 | 0.01 |
ENSDART00000137977
|
pear1
|
platelet endothelial aggregation receptor 1 |
| chr16_-_30564319 | 0.01 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr21_+_41649336 | 0.01 |
ENSDART00000164694
ENSDART00000181539 |
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr2_-_56655769 | 0.01 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr3_+_32698424 | 0.01 |
ENSDART00000055340
|
fus
|
FUS RNA binding protein |
| chr6_-_43005901 | 0.01 |
ENSDART00000161071
|
si:ch73-361p23.3
|
si:ch73-361p23.3 |
| chr9_+_29548195 | 0.01 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr25_-_19374710 | 0.01 |
ENSDART00000184483
ENSDART00000188706 |
map1ab
|
microtubule-associated protein 1Ab |
| chr11_-_3574311 | 0.01 |
ENSDART00000167064
|
si:dkey-33m11.7
|
si:dkey-33m11.7 |
| chr1_+_56447107 | 0.01 |
ENSDART00000091924
|
CABZ01059392.1
|
|
| chr8_-_2602572 | 0.01 |
ENSDART00000110482
|
zdhhc12a
|
zinc finger, DHHC-type containing 12a |
| chr11_+_28476298 | 0.01 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr12_+_16106459 | 0.01 |
ENSDART00000114174
|
muc13b
|
mucin 13b, cell surface associated |
| chr16_-_47426482 | 0.01 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr21_-_40557281 | 0.01 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr10_-_39321367 | 0.00 |
ENSDART00000129647
|
smtlb
|
somatolactin beta |
| chr16_-_24561354 | 0.00 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr1_+_8521323 | 0.00 |
ENSDART00000121439
ENSDART00000103626 ENSDART00000141283 |
mief2
|
mitochondrial elongation factor 2 |
| chr25_-_32888115 | 0.00 |
ENSDART00000087586
|
c2cd4a
|
C2 calcium dependent domain containing 4A |
| chr9_-_48937089 | 0.00 |
ENSDART00000193442
|
cers6
|
ceramide synthase 6 |
| chr1_-_12393060 | 0.00 |
ENSDART00000079782
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr1_-_21287724 | 0.00 |
ENSDART00000193900
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr2_-_8648440 | 0.00 |
ENSDART00000135743
|
si:ch211-71m22.3
|
si:ch211-71m22.3 |
| chr25_-_17918536 | 0.00 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of rorc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.1 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |