Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
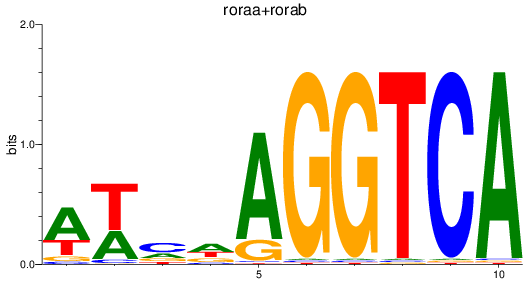
Results for roraa+rorab
Z-value: 0.92
Transcription factors associated with roraa+rorab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rorab
|
ENSDARG00000001910 | RAR-related orphan receptor A, paralog b |
|
roraa
|
ENSDARG00000031768 | RAR-related orphan receptor A, paralog a |
|
rorab
|
ENSDARG00000109324 | RAR-related orphan receptor A, paralog b |
|
rorab
|
ENSDARG00000113275 | RAR-related orphan receptor A, paralog b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rorab | dr11_v1_chr7_-_29571615_29571615 | 0.73 | 2.5e-02 | Click! |
| roraa | dr11_v1_chr25_+_33939728_33939728 | 0.47 | 2.0e-01 | Click! |
Activity profile of roraa+rorab motif
Sorted Z-values of roraa+rorab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_29900546 | 1.63 |
ENSDART00000147441
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr11_+_30729745 | 1.55 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr23_+_44741500 | 1.30 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr21_+_7582036 | 1.28 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr21_+_20771082 | 1.15 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr1_+_12763920 | 1.12 |
ENSDART00000189465
|
pcdh10a
|
protocadherin 10a |
| chr19_-_5332784 | 1.12 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr23_-_24488696 | 1.10 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr20_+_16750177 | 1.09 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr6_+_3710865 | 1.04 |
ENSDART00000170781
|
phospho2
|
phosphatase, orphan 2 |
| chr6_-_39764995 | 0.99 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr13_-_31452516 | 0.99 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr4_+_7677318 | 0.97 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr9_-_1970071 | 0.92 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr25_-_31396479 | 0.89 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr1_-_50859053 | 0.89 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr7_+_40228422 | 0.88 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr5_-_41831646 | 0.88 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr5_-_68916455 | 0.87 |
ENSDART00000171465
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr3_-_26109322 | 0.86 |
ENSDART00000113780
|
zgc:162612
|
zgc:162612 |
| chr16_-_28856112 | 0.86 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr15_+_19797918 | 0.85 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr10_+_37927100 | 0.84 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr7_+_29133321 | 0.81 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr5_+_20693724 | 0.81 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr19_+_40856534 | 0.80 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr4_-_1360495 | 0.77 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr8_+_25902170 | 0.76 |
ENSDART00000193130
|
rhoab
|
ras homolog gene family, member Ab |
| chr19_+_30662529 | 0.76 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr4_+_18963822 | 0.74 |
ENSDART00000066975
ENSDART00000066973 |
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr3_+_1182315 | 0.72 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr6_-_39765546 | 0.70 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_+_40884012 | 0.70 |
ENSDART00000149395
|
shha
|
sonic hedgehog a |
| chr7_-_4036184 | 0.67 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr18_+_8340886 | 0.67 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr10_+_20128267 | 0.67 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr5_+_1219187 | 0.67 |
ENSDART00000129490
|
gb:bc139872
|
expressed sequence BC139872 |
| chr10_-_29101715 | 0.66 |
ENSDART00000149674
ENSDART00000171194 ENSDART00000192019 |
si:ch211-103f14.3
|
si:ch211-103f14.3 |
| chr4_-_17629444 | 0.66 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr7_+_48805534 | 0.65 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr15_-_12270857 | 0.65 |
ENSDART00000170093
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr6_-_10835849 | 0.64 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr10_+_17850934 | 0.63 |
ENSDART00000113666
ENSDART00000145936 |
phf24
|
PHD finger protein 24 |
| chr20_+_40150612 | 0.61 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr7_-_24046999 | 0.61 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr19_+_40856807 | 0.59 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr14_+_33882973 | 0.58 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr23_-_15284757 | 0.57 |
ENSDART00000139135
|
sulf2b
|
sulfatase 2b |
| chr10_-_17103651 | 0.57 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr14_-_33454595 | 0.56 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr23_-_5683147 | 0.56 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr15_-_21239416 | 0.54 |
ENSDART00000155787
|
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr16_+_21242491 | 0.54 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr1_+_23783349 | 0.53 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr5_-_55395964 | 0.53 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr18_-_48492951 | 0.53 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr19_-_32487469 | 0.52 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr16_+_5678071 | 0.51 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr21_+_5882300 | 0.51 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr11_+_5842632 | 0.51 |
ENSDART00000111374
ENSDART00000158599 |
ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, (NADH-coenzyme Q reductase) |
| chr4_+_2620751 | 0.50 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr7_-_33351485 | 0.49 |
ENSDART00000146420
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr10_-_11385155 | 0.48 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr5_-_41838354 | 0.47 |
ENSDART00000146793
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr8_+_43016714 | 0.47 |
ENSDART00000142671
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr17_+_25290136 | 0.46 |
ENSDART00000173295
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr6_-_40312091 | 0.45 |
ENSDART00000154878
|
col7a1
|
collagen, type VII, alpha 1 |
| chr6_-_11523987 | 0.45 |
ENSDART00000189363
|
gulp1b
|
GULP, engulfment adaptor PTB domain containing 1b |
| chr10_+_20113830 | 0.45 |
ENSDART00000139722
|
dmtn
|
dematin actin binding protein |
| chr9_+_41914378 | 0.45 |
ENSDART00000130434
ENSDART00000007058 |
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr6_-_28980756 | 0.45 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr9_-_21067971 | 0.45 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr11_+_24900123 | 0.43 |
ENSDART00000044987
ENSDART00000148023 |
timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr16_+_12236339 | 0.43 |
ENSDART00000132468
|
tpi1b
|
triosephosphate isomerase 1b |
| chr12_-_45876387 | 0.42 |
ENSDART00000043210
ENSDART00000149044 |
pax2b
|
paired box 2b |
| chr7_+_48805725 | 0.41 |
ENSDART00000166543
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr10_-_7988396 | 0.41 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr7_+_20475788 | 0.41 |
ENSDART00000171155
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr14_+_9583588 | 0.41 |
ENSDART00000164101
|
tmem129
|
transmembrane protein 129, E3 ubiquitin protein ligase |
| chr2_-_56655769 | 0.41 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr1_-_45177373 | 0.40 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr18_-_1185772 | 0.40 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr22_-_24285432 | 0.39 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr20_-_25709247 | 0.39 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr8_+_22478090 | 0.39 |
ENSDART00000170263
|
si:ch211-261n11.7
|
si:ch211-261n11.7 |
| chr3_+_32698424 | 0.39 |
ENSDART00000055340
|
fus
|
FUS RNA binding protein |
| chr16_-_50229193 | 0.39 |
ENSDART00000161782
ENSDART00000010081 |
etfb
|
electron-transfer-flavoprotein, beta polypeptide |
| chr23_+_42813415 | 0.38 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr19_+_37620342 | 0.37 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr13_+_50375800 | 0.37 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr25_+_3326885 | 0.37 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr25_+_3327071 | 0.37 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr10_-_32524771 | 0.37 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr22_-_11124419 | 0.37 |
ENSDART00000149634
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr2_+_30787128 | 0.36 |
ENSDART00000189233
|
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr17_+_33340675 | 0.36 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr18_-_48517040 | 0.36 |
ENSDART00000143645
|
kcnj1a.3
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 3 |
| chr8_+_22477922 | 0.35 |
ENSDART00000189766
ENSDART00000179717 |
si:ch211-261n11.7
|
si:ch211-261n11.7 |
| chr1_+_34496855 | 0.34 |
ENSDART00000012873
|
klf12a
|
Kruppel-like factor 12a |
| chr23_-_19953089 | 0.33 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr6_-_1514767 | 0.32 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr10_+_1638876 | 0.31 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr22_+_21252790 | 0.31 |
ENSDART00000079046
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr14_-_29858883 | 0.31 |
ENSDART00000141034
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr2_-_4797512 | 0.31 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr9_-_1986014 | 0.31 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr3_+_12484008 | 0.31 |
ENSDART00000182229
|
vasnb
|
vasorin b |
| chr15_-_14486534 | 0.30 |
ENSDART00000179368
|
numbl
|
numb homolog (Drosophila)-like |
| chr4_-_25181552 | 0.30 |
ENSDART00000066930
|
atp5f1c
|
ATP synthase F1 subunit gamma |
| chr19_+_19976990 | 0.30 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr22_-_17574511 | 0.29 |
ENSDART00000181496
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr6_+_8339298 | 0.28 |
ENSDART00000151672
|
si:ch211-276a17.5
|
si:ch211-276a17.5 |
| chr5_-_41841892 | 0.27 |
ENSDART00000167089
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr3_+_58167288 | 0.27 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
| chr7_-_29341233 | 0.26 |
ENSDART00000140938
ENSDART00000147251 |
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr11_+_28476298 | 0.26 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr8_+_26125218 | 0.26 |
ENSDART00000145095
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr8_+_14158021 | 0.26 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr10_-_26273629 | 0.25 |
ENSDART00000147790
|
dchs1b
|
dachsous cadherin-related 1b |
| chr14_+_23518110 | 0.25 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr13_-_18195942 | 0.24 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr5_-_29780752 | 0.24 |
ENSDART00000137400
ENSDART00000145021 |
cfap77
|
cilia and flagella associated protein 77 |
| chr14_+_48045193 | 0.23 |
ENSDART00000124773
|
ppid
|
peptidylprolyl isomerase D |
| chr12_-_25201576 | 0.23 |
ENSDART00000077188
|
cox7a3
|
cytochrome c oxidase subunit VIIa polypeptide 3 |
| chr7_+_44484853 | 0.23 |
ENSDART00000189079
ENSDART00000121826 |
bean1
|
brain expressed, associated with NEDD4, 1 |
| chr15_+_45575180 | 0.23 |
ENSDART00000180441
|
cldn15lb
|
claudin 15-like b |
| chr10_+_4924388 | 0.22 |
ENSDART00000108595
|
slc46a2
|
solute carrier family 46 member 2 |
| chr6_+_51773873 | 0.22 |
ENSDART00000156516
|
tmem74b
|
transmembrane protein 74B |
| chr20_+_5564042 | 0.22 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr1_-_44704261 | 0.22 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr18_+_14529005 | 0.22 |
ENSDART00000186379
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr12_-_47648538 | 0.22 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr7_-_24005268 | 0.21 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr3_-_19091024 | 0.21 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr7_-_30227878 | 0.21 |
ENSDART00000173514
|
znf710b
|
zinc finger protein 710b |
| chr17_-_53353653 | 0.21 |
ENSDART00000180744
ENSDART00000026879 |
unm_sa911
|
un-named sa911 |
| chr12_-_45875946 | 0.21 |
ENSDART00000149970
|
pax2b
|
paired box 2b |
| chr2_+_58841181 | 0.21 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr21_+_15709061 | 0.20 |
ENSDART00000065772
|
ddt
|
D-dopachrome tautomerase |
| chr8_+_51084993 | 0.20 |
ENSDART00000127709
ENSDART00000053768 |
DISP3
|
si:dkey-32e23.6 |
| chr5_+_5689476 | 0.18 |
ENSDART00000022729
|
unm_sa808
|
un-named sa808 |
| chr18_+_36631923 | 0.18 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr15_+_29362714 | 0.18 |
ENSDART00000099916
ENSDART00000126196 |
gucy2f
|
guanylate cyclase 2F, retinal |
| chr6_-_49873020 | 0.17 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr12_+_7399403 | 0.17 |
ENSDART00000103526
|
bicc1b
|
BicC family RNA binding protein 1b |
| chr1_+_54115839 | 0.17 |
ENSDART00000180214
|
LO017722.2
|
|
| chr25_-_1235457 | 0.17 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr6_-_10320676 | 0.17 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr19_+_10331325 | 0.17 |
ENSDART00000143930
|
tmem238a
|
transmembrane protein 238a |
| chr10_+_19528321 | 0.16 |
ENSDART00000184816
|
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr17_-_26610814 | 0.16 |
ENSDART00000133402
ENSDART00000016608 |
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr11_-_37995501 | 0.16 |
ENSDART00000192096
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr7_-_8156169 | 0.15 |
ENSDART00000182960
ENSDART00000190184 ENSDART00000190885 ENSDART00000189867 ENSDART00000160836 |
si:cabz01030277.1
si:ch211-163c2.3
|
si:cabz01030277.1 si:ch211-163c2.3 |
| chr1_+_31573225 | 0.15 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr7_-_8014186 | 0.15 |
ENSDART00000190012
|
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr6_-_49170390 | 0.15 |
ENSDART00000182255
|
ngfb
|
nerve growth factor b (beta polypeptide) |
| chr19_+_20724347 | 0.15 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr1_+_44826593 | 0.15 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr1_+_31725154 | 0.14 |
ENSDART00000112333
ENSDART00000189801 |
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr14_-_30490465 | 0.14 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr24_-_21913426 | 0.14 |
ENSDART00000081178
|
c1qtnf9
|
C1q and TNF related 9 |
| chr2_-_16380283 | 0.13 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr25_+_31264155 | 0.13 |
ENSDART00000012256
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr14_-_29859067 | 0.13 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr6_-_9922266 | 0.13 |
ENSDART00000151549
|
pimr73
|
Pim proto-oncogene, serine/threonine kinase, related 73 |
| chr8_+_18463797 | 0.12 |
ENSDART00000148958
|
alkal1
|
ALK and LTK ligand 1 |
| chr4_+_9536860 | 0.11 |
ENSDART00000130083
|
lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr19_+_2590182 | 0.11 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr9_-_16877456 | 0.11 |
ENSDART00000161105
ENSDART00000160869 |
fbxl3a
|
F-box and leucine-rich repeat protein 3a |
| chr13_-_29406534 | 0.11 |
ENSDART00000100877
|
zgc:153142
|
zgc:153142 |
| chr22_+_29135846 | 0.11 |
ENSDART00000143811
|
baiap2l2b
|
BAI1-associated protein 2-like 2b |
| chr23_+_19564392 | 0.11 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr8_+_14792830 | 0.11 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr6_-_49159207 | 0.11 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr7_-_26446726 | 0.11 |
ENSDART00000173607
|
zgc:172079
|
zgc:172079 |
| chr9_-_53666031 | 0.10 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr1_+_19515228 | 0.10 |
ENSDART00000103091
|
clrn2
|
clarin 2 |
| chr21_-_22724980 | 0.10 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr15_+_1705167 | 0.10 |
ENSDART00000081940
|
otol1b
|
otolin 1b |
| chr12_-_44199316 | 0.10 |
ENSDART00000170378
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr8_+_30664077 | 0.10 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr17_-_36818176 | 0.09 |
ENSDART00000061762
|
myo6b
|
myosin VIb |
| chr17_+_15534815 | 0.09 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr16_+_26738404 | 0.09 |
ENSDART00000163110
|
fsbp
|
fibrinogen silencer binding protein |
| chr8_+_18464235 | 0.08 |
ENSDART00000110571
|
alkal1
|
ALK and LTK ligand 1 |
| chr18_+_7594012 | 0.08 |
ENSDART00000062150
|
zgc:77752
|
zgc:77752 |
| chr20_+_43623064 | 0.08 |
ENSDART00000186659
|
si:dkey-206f10.1
|
si:dkey-206f10.1 |
| chr16_-_16619854 | 0.08 |
ENSDART00000150512
ENSDART00000191306 ENSDART00000181773 ENSDART00000183231 |
cyp21a2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr11_-_29650930 | 0.08 |
ENSDART00000166969
|
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr16_+_27349585 | 0.07 |
ENSDART00000142573
|
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr19_-_3574060 | 0.07 |
ENSDART00000105120
|
tmem170b
|
transmembrane protein 170B |
| chr24_+_29449690 | 0.07 |
ENSDART00000105743
ENSDART00000193556 ENSDART00000145816 |
ntng1a
|
netrin g1a |
| chr2_-_44971551 | 0.07 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr15_+_31820536 | 0.07 |
ENSDART00000045921
|
frya
|
furry homolog a (Drosophila) |
| chr3_+_13637383 | 0.06 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr11_+_25278772 | 0.06 |
ENSDART00000188630
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr21_-_20840714 | 0.06 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr18_-_29962234 | 0.06 |
ENSDART00000144996
|
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr6_-_15603675 | 0.06 |
ENSDART00000143502
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr7_+_67494107 | 0.06 |
ENSDART00000185653
|
cpne7
|
copine VII |
Network of associatons between targets according to the STRING database.
First level regulatory network of roraa+rorab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 1.3 | GO:0051703 | mammillary body development(GO:0021767) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 0.5 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 0.7 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.2 | 1.7 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.6 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.5 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 0.4 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.8 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 1.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.4 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.3 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.1 | 0.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.3 | GO:0044060 | regulation of endocrine process(GO:0044060) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) endocrine hormone secretion(GO:0060986) |
| 0.1 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.3 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 1.1 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.4 | GO:0061075 | eye pigmentation(GO:0048069) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.5 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.7 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 1.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.2 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.9 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.5 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.9 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.5 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.5 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.1 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.4 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.8 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.4 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 1.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 1.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.4 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.1 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 1.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 1.0 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.5 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 1.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.4 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.1 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.4 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.6 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.5 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.2 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.4 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |