Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
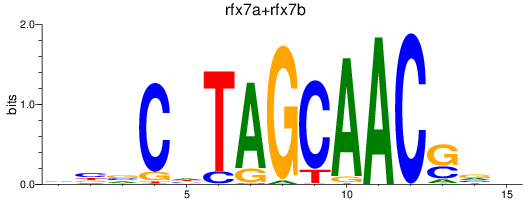
Results for rfx7a+rfx7b
Z-value: 1.08
Transcription factors associated with rfx7a+rfx7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rfx7a
|
ENSDARG00000077237 | regulatory factor X7a |
|
rfx7b
|
ENSDARG00000103691 | regulatory factor X7b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rfx7a | dr11_v1_chr7_+_34506937_34506937 | -0.39 | 3.0e-01 | Click! |
| rfx7b | dr11_v1_chr25_+_388258_388258 | -0.13 | 7.3e-01 | Click! |
Activity profile of rfx7a+rfx7b motif
Sorted Z-values of rfx7a+rfx7b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_36690742 | 1.17 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr24_-_9997948 | 0.62 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr24_-_10006158 | 0.56 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr18_+_2168695 | 0.55 |
ENSDART00000165855
|
dnaaf4
|
dynein axonemal assembly factor 4 |
| chr19_-_27588842 | 0.50 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr25_+_21895182 | 0.45 |
ENSDART00000152075
|
si:ch211-147k9.8
|
si:ch211-147k9.8 |
| chr3_+_21189766 | 0.43 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr20_-_3915770 | 0.42 |
ENSDART00000159322
|
si:ch73-111k22.3
|
si:ch73-111k22.3 |
| chr2_-_42960353 | 0.41 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr8_-_53488832 | 0.38 |
ENSDART00000191801
|
chdh
|
choline dehydrogenase |
| chr22_+_438714 | 0.36 |
ENSDART00000136491
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr20_+_30797329 | 0.34 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr4_-_12795436 | 0.34 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr24_+_22021675 | 0.33 |
ENSDART00000081234
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr25_-_10571078 | 0.32 |
ENSDART00000153898
|
si:ch211-107e6.5
|
si:ch211-107e6.5 |
| chr2_+_7075220 | 0.30 |
ENSDART00000022963
|
cdc14aa
|
cell division cycle 14Aa |
| chr6_+_4160579 | 0.30 |
ENSDART00000105278
ENSDART00000187932 ENSDART00000111817 |
trim25l
|
tripartite motif containing 25, like |
| chr14_-_35892767 | 0.30 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr7_-_24236364 | 0.30 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr21_-_29991098 | 0.29 |
ENSDART00000155772
|
si:ch73-190f9.4
|
si:ch73-190f9.4 |
| chr10_+_25204626 | 0.29 |
ENSDART00000024514
|
chordc1b
|
cysteine and histidine-rich domain (CHORD) containing 1b |
| chr23_+_24501918 | 0.28 |
ENSDART00000078824
|
szrd1
|
SUZ RNA binding domain containing 1 |
| chr20_-_7080427 | 0.28 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr14_+_24840669 | 0.28 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr25_+_35942867 | 0.27 |
ENSDART00000066985
|
hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr12_-_6159545 | 0.27 |
ENSDART00000152487
|
rltgr
|
RAMP-like triterpene glycoside receptor |
| chr24_-_36910224 | 0.25 |
ENSDART00000079233
|
dnaaf3l
|
dynein, axonemal, assembly factor 3 like |
| chr25_-_16755340 | 0.25 |
ENSDART00000124729
ENSDART00000110859 |
ribc2
|
RIB43A domain with coiled-coils 2 |
| chr1_-_51710225 | 0.25 |
ENSDART00000057601
ENSDART00000152745 |
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr15_-_47812268 | 0.24 |
ENSDART00000190231
|
tomt
|
transmembrane O-methyltransferase |
| chr4_-_78026285 | 0.24 |
ENSDART00000168273
|
CCT2
|
chaperonin containing TCP1 subunit 2 |
| chr13_+_24717880 | 0.24 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr18_-_5248365 | 0.24 |
ENSDART00000082506
ENSDART00000082504 ENSDART00000097960 |
myef2
|
myelin expression factor 2 |
| chr10_-_17231917 | 0.24 |
ENSDART00000038577
|
rab36
|
RAB36, member RAS oncogene family |
| chr6_-_40842768 | 0.23 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr24_+_22022109 | 0.23 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr13_+_24679674 | 0.23 |
ENSDART00000033090
ENSDART00000139854 |
zgc:66426
|
zgc:66426 |
| chr21_-_29990574 | 0.23 |
ENSDART00000185800
|
si:ch73-190f9.4
|
si:ch73-190f9.4 |
| chr11_-_40519886 | 0.23 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr21_-_29416966 | 0.23 |
ENSDART00000162575
|
zgc:171310
|
zgc:171310 |
| chr11_-_30352333 | 0.23 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr8_+_2487250 | 0.23 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr14_-_12051610 | 0.23 |
ENSDART00000193741
ENSDART00000029366 |
zgc:66447
|
zgc:66447 |
| chr8_-_49207319 | 0.23 |
ENSDART00000022870
|
fam110a
|
family with sequence similarity 110, member A |
| chr7_+_51795667 | 0.22 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr6_-_55297274 | 0.22 |
ENSDART00000184283
|
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr23_-_29357764 | 0.22 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr4_-_4261673 | 0.22 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr12_-_11649690 | 0.21 |
ENSDART00000149713
|
btbd16
|
BTB (POZ) domain containing 16 |
| chr21_-_29295713 | 0.21 |
ENSDART00000170081
|
CR318595.1
|
|
| chr7_+_17907883 | 0.21 |
ENSDART00000147564
|
mta2
|
metastasis associated 1 family, member 2 |
| chr12_+_19188542 | 0.21 |
ENSDART00000134726
ENSDART00000148011 ENSDART00000109541 |
cby1
|
chibby homolog 1 (Drosophila) |
| chr25_+_34845115 | 0.21 |
ENSDART00000061996
|
tmem231
|
transmembrane protein 231 |
| chr17_+_24684778 | 0.21 |
ENSDART00000146309
ENSDART00000082237 |
znf593
|
zinc finger protein 593 |
| chr17_-_10315644 | 0.21 |
ENSDART00000168572
|
ttc6
|
tetratricopeptide repeat domain 6 |
| chr1_-_59216197 | 0.21 |
ENSDART00000062426
|
lpar2b
|
lysophosphatidic acid receptor 2b |
| chr21_-_29940453 | 0.21 |
ENSDART00000182862
|
CU104716.1
|
|
| chr21_-_29838117 | 0.20 |
ENSDART00000160727
|
CR762480.1
|
|
| chr20_+_21022641 | 0.20 |
ENSDART00000152561
ENSDART00000141591 |
si:dkey-219e20.2
|
si:dkey-219e20.2 |
| chr21_-_29735366 | 0.20 |
ENSDART00000181668
|
CR847571.2
|
|
| chr7_+_17908235 | 0.20 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr20_-_52902693 | 0.20 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr14_-_33481428 | 0.20 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr13_-_18195942 | 0.20 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr3_-_31619463 | 0.20 |
ENSDART00000124559
|
moto
|
minamoto |
| chr25_+_3868438 | 0.19 |
ENSDART00000156438
|
tmem138
|
transmembrane protein 138 |
| chr4_-_14207471 | 0.19 |
ENSDART00000015134
|
twf1b
|
twinfilin actin-binding protein 1b |
| chr13_-_24717365 | 0.19 |
ENSDART00000137934
ENSDART00000003922 |
erlin1
|
ER lipid raft associated 1 |
| chr9_-_23156908 | 0.19 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr7_+_41146560 | 0.19 |
ENSDART00000143285
ENSDART00000173852 ENSDART00000174003 ENSDART00000038487 ENSDART00000173463 ENSDART00000166448 ENSDART00000052274 |
puf60b
|
poly-U binding splicing factor b |
| chr5_+_25681174 | 0.19 |
ENSDART00000134773
|
zfand5a
|
zinc finger, AN1-type domain 5a |
| chr7_+_22982201 | 0.18 |
ENSDART00000134116
|
ccnb3
|
cyclin B3 |
| chr21_-_29364166 | 0.18 |
ENSDART00000188602
|
BX537120.3
|
|
| chr12_-_48168135 | 0.18 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr8_+_31016180 | 0.18 |
ENSDART00000130870
ENSDART00000143604 |
odf2b
|
outer dense fiber of sperm tails 2b |
| chr10_-_17232372 | 0.18 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr1_+_58609885 | 0.18 |
ENSDART00000186199
|
si:ch73-236c18.9
|
si:ch73-236c18.9 |
| chr3_-_60175470 | 0.18 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr25_+_1035656 | 0.18 |
ENSDART00000179843
ENSDART00000168637 |
si:ch211-284k5.2
|
si:ch211-284k5.2 |
| chr14_-_38827442 | 0.17 |
ENSDART00000160000
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr20_+_33519435 | 0.17 |
ENSDART00000061829
|
drc1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr13_-_6323959 | 0.17 |
ENSDART00000149619
ENSDART00000148907 |
mcph1
|
microcephalin 1 |
| chr12_+_17620067 | 0.17 |
ENSDART00000073599
|
rsph10b
|
radial spoke head 10 homolog B |
| chr11_+_37652870 | 0.17 |
ENSDART00000129918
|
kif17
|
kinesin family member 17 |
| chr25_-_28638052 | 0.17 |
ENSDART00000138918
ENSDART00000135247 ENSDART00000114662 ENSDART00000157493 ENSDART00000137677 |
agbl2
|
ATP/GTP binding protein-like 2 |
| chr10_+_44692272 | 0.17 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr21_-_32781612 | 0.17 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr25_+_34845469 | 0.17 |
ENSDART00000145416
|
tmem231
|
transmembrane protein 231 |
| chr6_-_19683406 | 0.16 |
ENSDART00000158041
|
cfap52
|
cilia and flagella associated protein 52 |
| chr17_+_11500628 | 0.16 |
ENSDART00000155660
|
efcab2
|
EF-hand calcium binding domain 2 |
| chr1_-_6225285 | 0.16 |
ENSDART00000141653
|
mdh1b
|
malate dehydrogenase 1B, NAD (soluble) |
| chr21_-_29533723 | 0.16 |
ENSDART00000193554
|
BX537120.2
|
|
| chr13_+_159504 | 0.16 |
ENSDART00000057248
|
ewsr1b
|
EWS RNA-binding protein 1b |
| chr1_+_45002971 | 0.16 |
ENSDART00000021336
|
dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr10_-_10863936 | 0.16 |
ENSDART00000180568
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr13_+_233482 | 0.16 |
ENSDART00000102511
|
cfap36
|
cilia and flagella associated protein 36 |
| chr2_+_16449627 | 0.16 |
ENSDART00000005381
|
zgc:110269
|
zgc:110269 |
| chr14_-_30905288 | 0.16 |
ENSDART00000173449
ENSDART00000173451 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr5_+_36650096 | 0.16 |
ENSDART00000111414
|
alkbh6
|
alkB homolog 6 |
| chr5_+_39563301 | 0.15 |
ENSDART00000131245
ENSDART00000097872 |
si:ch211-57h10.1
|
si:ch211-57h10.1 |
| chr8_-_16515127 | 0.15 |
ENSDART00000146469
ENSDART00000132681 |
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr21_-_29689141 | 0.15 |
ENSDART00000100876
|
CR847571.1
|
|
| chr23_+_12134839 | 0.15 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr17_-_46933567 | 0.15 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr13_-_24717618 | 0.15 |
ENSDART00000172156
|
erlin1
|
ER lipid raft associated 1 |
| chr5_-_57920600 | 0.15 |
ENSDART00000132376
|
si:dkey-27p23.3
|
si:dkey-27p23.3 |
| chr10_-_15854743 | 0.14 |
ENSDART00000092343
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr18_-_38270077 | 0.14 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr14_-_38826739 | 0.14 |
ENSDART00000187633
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr17_-_18888959 | 0.14 |
ENSDART00000080029
|
ak7b
|
adenylate kinase 7b |
| chr5_-_66823750 | 0.14 |
ENSDART00000041441
ENSDART00000112488 |
stip1
|
stress-induced phosphoprotein 1 |
| chr4_+_34343524 | 0.14 |
ENSDART00000171694
|
si:ch211-246b8.5
|
si:ch211-246b8.5 |
| chr3_+_60034194 | 0.14 |
ENSDART00000189858
ENSDART00000153642 |
CR352258.1
|
|
| chr8_+_45293743 | 0.14 |
ENSDART00000136234
ENSDART00000137104 |
ubap2b
|
ubiquitin associated protein 2b |
| chr1_-_53277187 | 0.14 |
ENSDART00000131520
|
znf330
|
zinc finger protein 330 |
| chr15_-_23206562 | 0.14 |
ENSDART00000110360
|
ccdc153
|
coiled-coil domain containing 153 |
| chr7_+_16047863 | 0.14 |
ENSDART00000188750
|
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr17_+_27545183 | 0.14 |
ENSDART00000129392
|
pacrg
|
PARK2 co-regulated |
| chr11_+_3585934 | 0.14 |
ENSDART00000055694
|
cdab
|
cytidine deaminase b |
| chr10_-_10864331 | 0.13 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr5_-_24882357 | 0.13 |
ENSDART00000127773
ENSDART00000003998 ENSDART00000143851 |
ewsr1b
|
EWS RNA-binding protein 1b |
| chr16_-_31791165 | 0.13 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr21_-_23307653 | 0.13 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr13_-_48764180 | 0.13 |
ENSDART00000167157
|
si:ch1073-266p11.2
|
si:ch1073-266p11.2 |
| chr4_-_71110826 | 0.13 |
ENSDART00000167431
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr11_-_34720629 | 0.13 |
ENSDART00000162066
ENSDART00000161471 |
dnah1
|
dynein, axonemal, heavy chain 1 |
| chr7_-_27037990 | 0.12 |
ENSDART00000173561
|
nucb2a
|
nucleobindin 2a |
| chr17_+_53186347 | 0.12 |
ENSDART00000155981
|
dph6
|
diphthamine biosynthesis 6 |
| chr11_-_12232929 | 0.12 |
ENSDART00000127611
|
si:ch211-156l18.4
|
si:ch211-156l18.4 |
| chr11_-_12417952 | 0.12 |
ENSDART00000135694
|
si:dkey-27d5.9
|
si:dkey-27d5.9 |
| chr17_-_22552678 | 0.12 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr20_+_18225329 | 0.12 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr17_+_389218 | 0.12 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr17_-_48805159 | 0.12 |
ENSDART00000103662
|
kif6
|
kinesin family member 6 |
| chr21_-_45086170 | 0.12 |
ENSDART00000188963
|
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr11_+_25328199 | 0.12 |
ENSDART00000141478
ENSDART00000112209 |
fam83d
|
family with sequence similarity 83, member D |
| chr7_-_40738774 | 0.11 |
ENSDART00000084179
|
rnf32
|
ring finger protein 32 |
| chr3_-_12026741 | 0.11 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr7_-_22981796 | 0.11 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr14_+_30413312 | 0.11 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr3_+_60716904 | 0.11 |
ENSDART00000168280
|
foxj1a
|
forkhead box J1a |
| chr11_+_11387509 | 0.11 |
ENSDART00000159578
|
si:dkey-23f9.9
|
si:dkey-23f9.9 |
| chr9_+_48219111 | 0.11 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr15_+_29085955 | 0.11 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr11_-_12717004 | 0.11 |
ENSDART00000122020
|
si:ch211-209f22.3
|
si:ch211-209f22.3 |
| chr2_-_7246848 | 0.11 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr24_-_37272116 | 0.11 |
ENSDART00000022999
|
ube2ib
|
ubiquitin-conjugating enzyme E2Ib |
| chr9_+_54716436 | 0.11 |
ENSDART00000000552
|
ofd1
|
oral-facial-digital syndrome 1 |
| chr22_-_24984733 | 0.11 |
ENSDART00000142147
ENSDART00000187284 |
dnal4b
|
dynein, axonemal, light chain 4b |
| chr7_+_22981909 | 0.11 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr15_+_37589698 | 0.11 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr19_-_27395531 | 0.11 |
ENSDART00000103940
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr22_-_18546649 | 0.10 |
ENSDART00000171277
|
cirbpb
|
cold inducible RNA binding protein b |
| chr17_-_28811747 | 0.10 |
ENSDART00000001444
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr7_+_50464500 | 0.10 |
ENSDART00000191356
|
serinc4
|
serine incorporator 4 |
| chr7_+_8670398 | 0.10 |
ENSDART00000164195
|
si:ch211-1o7.2
|
si:ch211-1o7.2 |
| chr7_-_27038488 | 0.10 |
ENSDART00000052731
ENSDART00000191382 |
nucb2a
|
nucleobindin 2a |
| chr8_-_14080534 | 0.10 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr16_+_28578352 | 0.10 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr16_+_50006872 | 0.10 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr11_-_12437234 | 0.10 |
ENSDART00000169324
|
zgc:174355
|
zgc:174355 |
| chr17_-_49412313 | 0.10 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr13_-_3155243 | 0.10 |
ENSDART00000139183
ENSDART00000050934 |
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr11_-_12394682 | 0.10 |
ENSDART00000172447
|
si:dkey-27d5.10
|
si:dkey-27d5.10 |
| chr19_+_3849378 | 0.10 |
ENSDART00000166218
ENSDART00000159228 |
oscp1a
|
organic solute carrier partner 1a |
| chr11_-_12219829 | 0.10 |
ENSDART00000168025
ENSDART00000112677 ENSDART00000108681 |
si:ch211-156l18.6
|
si:ch211-156l18.6 |
| chr24_-_17067284 | 0.10 |
ENSDART00000111237
|
armc3
|
armadillo repeat containing 3 |
| chr22_-_37565348 | 0.09 |
ENSDART00000149482
ENSDART00000104478 |
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr24_+_26345609 | 0.09 |
ENSDART00000186844
|
lrrc34
|
leucine rich repeat containing 34 |
| chr23_+_37090889 | 0.09 |
ENSDART00000074406
|
ubxn10
|
UBX domain protein 10 |
| chr13_+_24842857 | 0.09 |
ENSDART00000123866
|
dusp13a
|
dual specificity phosphatase 13a |
| chr13_-_33246154 | 0.09 |
ENSDART00000130127
|
rtf1
|
RTF1 homolog, Paf1/RNA polymerase II complex component |
| chr24_+_36239617 | 0.09 |
ENSDART00000062745
|
riok3
|
RIO kinase 3 (yeast) |
| chr13_-_51903150 | 0.09 |
ENSDART00000090644
|
mrpl2
|
mitochondrial ribosomal protein L2 |
| chr11_+_11430552 | 0.09 |
ENSDART00000160339
|
si:dkey-23f9.11
|
si:dkey-23f9.11 |
| chr18_-_14677936 | 0.09 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr2_-_59247811 | 0.09 |
ENSDART00000141384
|
ftr32
|
finTRIM family, member 32 |
| chr22_+_25049563 | 0.09 |
ENSDART00000078173
|
dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr17_+_31592191 | 0.08 |
ENSDART00000153765
|
si:dkey-13p1.3
|
si:dkey-13p1.3 |
| chr7_+_38395197 | 0.08 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr7_+_26549846 | 0.08 |
ENSDART00000141353
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr22_+_30046365 | 0.08 |
ENSDART00000192546
|
add3a
|
adducin 3 (gamma) a |
| chr17_-_24684687 | 0.08 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr25_+_25438322 | 0.08 |
ENSDART00000150364
|
LRRC56
|
si:ch211-103e16.5 |
| chr7_-_29533127 | 0.08 |
ENSDART00000057289
|
anxa2b
|
annexin A2b |
| chr17_+_46935061 | 0.08 |
ENSDART00000156578
|
pimr26
|
Pim proto-oncogene, serine/threonine kinase, related 26 |
| chr24_+_26345427 | 0.08 |
ENSDART00000089756
|
lrrc34
|
leucine rich repeat containing 34 |
| chr7_+_7630409 | 0.08 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr13_-_36703164 | 0.08 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr11_-_12379541 | 0.08 |
ENSDART00000171717
|
zgc:174353
|
zgc:174353 |
| chr16_+_54641230 | 0.08 |
ENSDART00000157641
ENSDART00000159540 |
fbxo43
|
F-box protein 43 |
| chr24_+_14240196 | 0.08 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr11_+_11374021 | 0.08 |
ENSDART00000170806
|
si:dkey-23f9.8
|
si:dkey-23f9.8 |
| chr2_-_55337585 | 0.08 |
ENSDART00000177924
|
tpm4b
|
tropomyosin 4b |
| chr11_+_11360641 | 0.08 |
ENSDART00000159765
|
si:dkey-23f9.7
|
si:dkey-23f9.7 |
| chr21_-_45348983 | 0.07 |
ENSDART00000188017
|
skp1
|
S-phase kinase-associated protein 1 |
| chr17_-_33716688 | 0.07 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr12_+_28955766 | 0.07 |
ENSDART00000123417
ENSDART00000139347 |
znf668
|
zinc finger protein 668 |
| chr12_+_11650146 | 0.07 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr14_+_10596628 | 0.07 |
ENSDART00000115177
|
gpr174
|
G protein-coupled receptor 174 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx7a+rfx7b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 1.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.2 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.3 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.4 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.4 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.3 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.2 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0071922 | regulation of sister chromatid cohesion(GO:0007063) regulation of chromosome condensation(GO:0060623) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.3 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.2 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.1 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.6 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.0 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.3 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.3 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.0 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0051219 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.0 | 0.0 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |