Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
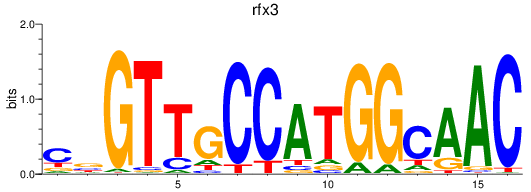
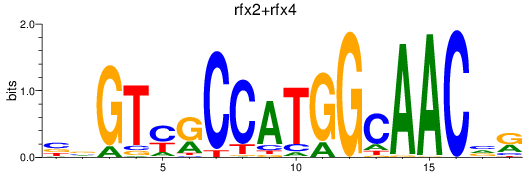
Results for rfx3_rfx2+rfx4
Z-value: 3.72
Transcription factors associated with rfx3_rfx2+rfx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rfx3
|
ENSDARG00000014550 | regulatory factor X, 3 (influences HLA class II expression) |
|
rfx2
|
ENSDARG00000013575 | regulatory factor X, 2 (influences HLA class II expression) |
|
rfx4
|
ENSDARG00000026395 | regulatory factor X, 4 |
|
rfx4
|
ENSDARG00000116861 | regulatory factor X, 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rfx4 | dr11_v1_chr18_-_15373620_15373620 | 0.95 | 6.8e-05 | Click! |
| rfx3 | dr11_v1_chr10_-_625441_625441 | -0.91 | 5.4e-04 | Click! |
| rfx2 | dr11_v1_chr8_-_20118549_20118549 | -0.87 | 2.6e-03 | Click! |
Activity profile of rfx3_rfx2+rfx4 motif
Sorted Z-values of rfx3_rfx2+rfx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_68789 | 22.05 |
ENSDART00000058569
|
cldn1
|
claudin 1 |
| chr9_-_712308 | 9.66 |
ENSDART00000144625
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr24_-_205275 | 7.87 |
ENSDART00000108762
|
vopp1
|
VOPP1, WBP1/VOPP1 family member |
| chr8_+_21195420 | 7.40 |
ENSDART00000100234
ENSDART00000091307 |
col2a1a
|
collagen, type II, alpha 1a |
| chr23_+_35708730 | 5.64 |
ENSDART00000009277
|
tuba1a
|
tubulin, alpha 1a |
| chr6_+_612330 | 5.45 |
ENSDART00000166872
ENSDART00000191758 |
kynu
|
kynureninase |
| chr16_-_45069882 | 5.38 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr21_+_26697536 | 5.32 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr5_-_71705191 | 4.67 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr6_-_8736766 | 4.63 |
ENSDART00000143956
|
cavin2b
|
caveolae associated protein 2b |
| chr21_-_15929041 | 4.48 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr7_-_18554603 | 4.39 |
ENSDART00000108938
|
msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr17_+_53186347 | 4.22 |
ENSDART00000155981
|
dph6
|
diphthamine biosynthesis 6 |
| chr19_-_10551394 | 4.12 |
ENSDART00000186815
ENSDART00000181041 |
lim2.4
|
lens intrinsic membrane protein 2.4 |
| chr13_-_31470439 | 4.05 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr7_-_34265481 | 3.82 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr3_-_1190132 | 3.75 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr11_-_279328 | 3.69 |
ENSDART00000066179
|
npffl
|
neuropeptide FF-amide peptide precursor like |
| chr7_+_1045637 | 3.54 |
ENSDART00000111531
|
epdl1
|
ependymin-like 1 |
| chr13_-_36703164 | 3.51 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr14_-_25577094 | 3.48 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr24_-_37680917 | 3.47 |
ENSDART00000131342
|
anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr17_-_21784152 | 3.47 |
ENSDART00000127254
|
hmx2
|
H6 family homeobox 2 |
| chr3_+_60716904 | 3.44 |
ENSDART00000168280
|
foxj1a
|
forkhead box J1a |
| chr5_+_39563301 | 3.28 |
ENSDART00000131245
ENSDART00000097872 |
si:ch211-57h10.1
|
si:ch211-57h10.1 |
| chr8_-_19216657 | 3.20 |
ENSDART00000135096
ENSDART00000135869 ENSDART00000145951 |
si:ch73-222f22.2
si:ch73-222f22.2
|
si:ch73-222f22.2 si:ch73-222f22.2 |
| chr13_+_33117528 | 3.12 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr18_+_30028135 | 3.07 |
ENSDART00000140825
ENSDART00000145306 ENSDART00000136810 |
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr15_+_47903864 | 2.99 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr17_-_10838434 | 2.98 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr3_-_19899914 | 2.93 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr2_+_35728033 | 2.93 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr11_+_43661735 | 2.88 |
ENSDART00000017912
|
gli2b
|
GLI family zinc finger 2b |
| chr25_+_37480285 | 2.87 |
ENSDART00000166187
|
CABZ01095001.1
|
|
| chr10_-_29900546 | 2.87 |
ENSDART00000147441
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr19_-_9472893 | 2.85 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr25_+_25453571 | 2.84 |
ENSDART00000112330
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr17_+_33767890 | 2.81 |
ENSDART00000193177
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr8_-_19216180 | 2.73 |
ENSDART00000146162
|
si:ch73-222f22.2
|
si:ch73-222f22.2 |
| chr18_+_30028637 | 2.72 |
ENSDART00000139750
|
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr16_+_50006872 | 2.68 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr24_-_38816725 | 2.63 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr16_+_24632096 | 2.58 |
ENSDART00000157237
|
si:dkey-56f14.7
|
si:dkey-56f14.7 |
| chr2_-_5728843 | 2.57 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr13_+_50368668 | 2.57 |
ENSDART00000127610
|
dnajc12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_-_470812 | 2.56 |
ENSDART00000192527
|
zgc:92518
|
zgc:92518 |
| chr20_-_13640598 | 2.55 |
ENSDART00000128823
ENSDART00000103394 |
rsph3
|
radial spoke 3 homolog |
| chr7_-_24236364 | 2.54 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr5_-_23179319 | 2.53 |
ENSDART00000161883
ENSDART00000136260 |
si:dkey-114c15.5
|
si:dkey-114c15.5 |
| chr17_-_6759006 | 2.50 |
ENSDART00000184692
ENSDART00000180530 |
vsnl1b
|
visinin-like 1b |
| chr8_-_54304381 | 2.48 |
ENSDART00000184177
|
RHO (1 of many)
|
rhodopsin |
| chr17_-_24684687 | 2.48 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr4_+_31259 | 2.46 |
ENSDART00000166826
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr4_-_5652030 | 2.46 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr21_-_5879897 | 2.46 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr19_+_22062202 | 2.43 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr24_-_24163201 | 2.37 |
ENSDART00000140170
|
map7d2b
|
MAP7 domain containing 2b |
| chr20_+_20638034 | 2.37 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr13_-_3155243 | 2.36 |
ENSDART00000139183
ENSDART00000050934 |
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr6_+_52212927 | 2.36 |
ENSDART00000143458
|
ywhaba
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a |
| chr11_-_44194132 | 2.35 |
ENSDART00000182954
ENSDART00000111271 |
CABZ01080074.1
|
|
| chr20_+_20637866 | 2.34 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr5_-_55625247 | 2.30 |
ENSDART00000133156
|
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr13_+_28821841 | 2.30 |
ENSDART00000179900
|
CU639469.1
|
|
| chr8_-_49345388 | 2.30 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr13_-_13294847 | 2.28 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr6_+_52212574 | 2.28 |
ENSDART00000025940
|
ywhaba
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a |
| chr1_+_54908895 | 2.28 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr20_-_9436521 | 2.27 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr8_+_23809843 | 2.26 |
ENSDART00000099724
|
si:ch211-163l21.10
|
si:ch211-163l21.10 |
| chr6_-_21582444 | 2.21 |
ENSDART00000151339
|
si:dkey-43k4.3
|
si:dkey-43k4.3 |
| chr16_-_24612871 | 2.21 |
ENSDART00000155614
ENSDART00000154787 ENSDART00000155983 ENSDART00000156519 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr10_-_17232372 | 2.20 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr3_-_50277959 | 2.17 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr23_-_20133994 | 2.15 |
ENSDART00000004871
|
lrrc23
|
leucine rich repeat containing 23 |
| chr10_-_36682509 | 2.14 |
ENSDART00000148093
ENSDART00000063365 |
dnajb13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr24_-_24162930 | 2.12 |
ENSDART00000080602
|
map7d2b
|
MAP7 domain containing 2b |
| chr22_-_25033105 | 2.12 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr7_-_58098814 | 2.12 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr12_+_25600685 | 2.10 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr25_+_25453221 | 2.08 |
ENSDART00000176822
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr10_-_43117607 | 2.06 |
ENSDART00000148293
ENSDART00000089965 |
tmem167a
|
transmembrane protein 167A |
| chr16_+_46492756 | 2.03 |
ENSDART00000105931
|
rpz5
|
rapunzel 5 |
| chr21_+_22423286 | 2.02 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr21_+_17956856 | 2.01 |
ENSDART00000080431
|
dnai1.2
|
dynein, axonemal, intermediate chain 1, paralog 2 |
| chr3_-_22829710 | 2.00 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr20_-_3915770 | 1.99 |
ENSDART00000159322
|
si:ch73-111k22.3
|
si:ch73-111k22.3 |
| chr2_-_56348727 | 1.98 |
ENSDART00000060745
|
uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr24_-_32408404 | 1.98 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr19_+_7567763 | 1.96 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr13_+_111212 | 1.96 |
ENSDART00000167840
|
dnaaf2
|
dynein, axonemal, assembly factor 2 |
| chr14_-_52542928 | 1.95 |
ENSDART00000075749
|
ppp2r2ba
|
protein phosphatase 2, regulatory subunit B, beta a |
| chr1_-_10806625 | 1.95 |
ENSDART00000139749
|
si:ch73-222h13.1
|
si:ch73-222h13.1 |
| chr4_-_390431 | 1.94 |
ENSDART00000067482
ENSDART00000138500 |
dynlt1
|
dynein, light chain, Tctex-type 1 |
| chr8_-_32803227 | 1.94 |
ENSDART00000110079
|
zgc:194839
|
zgc:194839 |
| chr25_+_13731542 | 1.94 |
ENSDART00000161012
|
ccdc135
|
coiled-coil domain containing 135 |
| chr5_-_57920600 | 1.91 |
ENSDART00000132376
|
si:dkey-27p23.3
|
si:dkey-27p23.3 |
| chr6_+_120181 | 1.87 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr9_-_3671911 | 1.86 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr7_+_41295974 | 1.85 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr1_-_45633955 | 1.85 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr15_-_19669770 | 1.84 |
ENSDART00000152707
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr3_-_61181018 | 1.83 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr21_-_17956416 | 1.83 |
ENSDART00000026737
|
stx2a
|
syntaxin 2a |
| chr21_-_22828593 | 1.83 |
ENSDART00000150993
|
angptl5
|
angiopoietin-like 5 |
| chr17_-_53440284 | 1.82 |
ENSDART00000126976
|
mycbp
|
c-myc binding protein |
| chr5_-_54672763 | 1.81 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr7_+_22675475 | 1.81 |
ENSDART00000134423
|
ponzr3
|
plac8 onzin related protein 3 |
| chr2_+_6126638 | 1.80 |
ENSDART00000153916
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr15_+_44053244 | 1.79 |
ENSDART00000059550
|
lrrc51
|
leucine rich repeat containing 51 |
| chr17_+_53250802 | 1.79 |
ENSDART00000143819
|
VASH1
|
vasohibin 1 |
| chr21_-_17956739 | 1.78 |
ENSDART00000148154
|
stx2a
|
syntaxin 2a |
| chr24_-_2917540 | 1.77 |
ENSDART00000164776
|
fam69c
|
family with sequence similarity 69, member C |
| chr3_-_6767440 | 1.76 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr6_+_13933464 | 1.75 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr21_+_41743493 | 1.75 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr22_-_16494406 | 1.71 |
ENSDART00000062727
|
stx6
|
syntaxin 6 |
| chr2_-_30659222 | 1.70 |
ENSDART00000145405
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr3_-_52644737 | 1.70 |
ENSDART00000126180
|
si:dkey-210j14.3
|
si:dkey-210j14.3 |
| chr10_+_37145007 | 1.68 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr10_-_29744921 | 1.68 |
ENSDART00000088605
|
ift46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr11_-_97817 | 1.67 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr18_+_46382484 | 1.67 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr14_-_36799280 | 1.66 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr10_-_35542071 | 1.63 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr18_+_34601023 | 1.61 |
ENSDART00000149647
|
tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr6_+_7250824 | 1.61 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr8_-_25338709 | 1.60 |
ENSDART00000131616
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr13_-_49819027 | 1.59 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr8_-_20380750 | 1.59 |
ENSDART00000113910
|
adamts10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| chr10_-_9410068 | 1.58 |
ENSDART00000041382
|
morn5
|
MORN repeat containing 5 |
| chr19_-_4023050 | 1.58 |
ENSDART00000162883
|
stpg1
|
sperm-tail PG-rich repeat containing 1 |
| chr23_+_43950674 | 1.58 |
ENSDART00000167813
|
corin
|
corin, serine peptidase |
| chr6_+_47424266 | 1.57 |
ENSDART00000171087
|
si:ch211-286o17.1
|
si:ch211-286o17.1 |
| chr1_-_56223913 | 1.56 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr10_-_22797959 | 1.55 |
ENSDART00000183269
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr16_+_221739 | 1.55 |
ENSDART00000180243
|
nrsn1
|
neurensin 1 |
| chr19_-_103289 | 1.54 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr2_-_14271874 | 1.53 |
ENSDART00000189390
|
tctex1d1
|
Tctex1 domain containing 1 |
| chr3_+_31945604 | 1.52 |
ENSDART00000114957
|
zgc:193811
|
zgc:193811 |
| chr15_-_31177324 | 1.51 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr20_-_32270866 | 1.49 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr9_-_327901 | 1.47 |
ENSDART00000159956
|
ndufa4l2a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2a |
| chr13_+_33246150 | 1.47 |
ENSDART00000136356
ENSDART00000132834 ENSDART00000144379 ENSDART00000131985 ENSDART00000148368 ENSDART00000035940 ENSDART00000141912 |
ndufaf1
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr16_-_50175069 | 1.42 |
ENSDART00000192979
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr6_-_33023745 | 1.42 |
ENSDART00000156211
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr8_-_53488832 | 1.42 |
ENSDART00000191801
|
chdh
|
choline dehydrogenase |
| chr22_+_2937485 | 1.41 |
ENSDART00000082222
ENSDART00000135507 ENSDART00000143258 |
cep19
|
centrosomal protein 19 |
| chr21_-_22709251 | 1.41 |
ENSDART00000140032
|
si:dkeyp-69c1.9
|
si:dkeyp-69c1.9 |
| chr24_+_17269849 | 1.40 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr15_-_20939579 | 1.40 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr5_-_42661012 | 1.40 |
ENSDART00000158339
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr2_-_14272083 | 1.39 |
ENSDART00000169986
|
tctex1d1
|
Tctex1 domain containing 1 |
| chr19_-_40186328 | 1.39 |
ENSDART00000087474
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr10_-_17231917 | 1.39 |
ENSDART00000038577
|
rab36
|
RAB36, member RAS oncogene family |
| chr4_+_77971104 | 1.39 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr6_+_3816241 | 1.38 |
ENSDART00000178545
|
erich2
|
glutamate-rich 2 |
| chr4_-_42242844 | 1.38 |
ENSDART00000163476
|
si:ch211-129p6.2
|
si:ch211-129p6.2 |
| chr20_+_52458765 | 1.38 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr16_+_46492994 | 1.38 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr25_+_13731726 | 1.38 |
ENSDART00000181065
|
ccdc135
|
coiled-coil domain containing 135 |
| chr25_-_28638052 | 1.37 |
ENSDART00000138918
ENSDART00000135247 ENSDART00000114662 ENSDART00000157493 ENSDART00000137677 |
agbl2
|
ATP/GTP binding protein-like 2 |
| chr20_+_27691307 | 1.36 |
ENSDART00000153299
|
si:dkey-1h6.8
|
si:dkey-1h6.8 |
| chr16_+_4497302 | 1.34 |
ENSDART00000081826
ENSDART00000148096 |
ttc29
|
tetratricopeptide repeat domain 29 |
| chr4_-_13626831 | 1.33 |
ENSDART00000173077
|
si:dkeyp-81f3.4
|
si:dkeyp-81f3.4 |
| chr4_+_3980247 | 1.32 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr10_+_9410304 | 1.31 |
ENSDART00000080843
|
ndufa8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
| chr1_+_37391716 | 1.30 |
ENSDART00000191986
|
sparcl1
|
SPARC-like 1 |
| chr2_+_47581997 | 1.29 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr25_-_25736958 | 1.29 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr19_+_5318358 | 1.29 |
ENSDART00000082133
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr17_-_39779906 | 1.29 |
ENSDART00000155181
|
pimr61
|
Pim proto-oncogene, serine/threonine kinase, related 61 |
| chr5_-_57652168 | 1.28 |
ENSDART00000166245
|
gb:eh456644
|
expressed sequence EH456644 |
| chr19_-_12324514 | 1.27 |
ENSDART00000122192
|
ncaldb
|
neurocalcin delta b |
| chr20_-_43775495 | 1.27 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr2_-_197665 | 1.27 |
ENSDART00000169743
|
CABZ01064859.1
|
|
| chr9_-_31747106 | 1.26 |
ENSDART00000048469
ENSDART00000145204 ENSDART00000186889 |
nalcn
|
sodium leak channel, non-selective |
| chr4_-_8611841 | 1.25 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr5_-_8406192 | 1.23 |
ENSDART00000159718
|
spef2
|
sperm flagellar 2 |
| chr19_-_1405363 | 1.22 |
ENSDART00000165516
|
cfap20
|
cilia and flagella associated protein 20 |
| chr17_-_53439866 | 1.21 |
ENSDART00000154826
|
mycbp
|
c-myc binding protein |
| chr17_-_40397752 | 1.21 |
ENSDART00000178483
|
BX548062.1
|
|
| chr11_-_11518469 | 1.20 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr1_+_10720294 | 1.18 |
ENSDART00000139387
|
atp1b1b
|
ATPase Na+/K+ transporting subunit beta 1b |
| chr11_+_36477481 | 1.17 |
ENSDART00000128245
|
ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr22_+_16418688 | 1.17 |
ENSDART00000009360
|
ankrd29
|
ankyrin repeat domain 29 |
| chr24_+_17270129 | 1.17 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr9_-_6927587 | 1.17 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr4_-_9586713 | 1.16 |
ENSDART00000145613
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr5_+_64732036 | 1.16 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr14_-_25042184 | 1.16 |
ENSDART00000131027
|
TMEM216
|
si:rp71-1d10.5 |
| chr8_+_26522013 | 1.15 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr9_+_9441453 | 1.14 |
ENSDART00000081859
ENSDART00000188567 ENSDART00000143101 |
maats1
|
MYCBP-associated, testis expressed 1 |
| chr21_+_39292266 | 1.14 |
ENSDART00000144116
|
si:ch211-274p24.2
|
si:ch211-274p24.2 |
| chr5_+_42400777 | 1.13 |
ENSDART00000183114
|
BX548073.8
|
|
| chr9_+_17309195 | 1.13 |
ENSDART00000048548
|
scel
|
sciellin |
| chr8_+_2487883 | 1.12 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr17_-_44249720 | 1.12 |
ENSDART00000156648
|
otx2b
|
orthodenticle homeobox 2b |
| chr15_+_33865312 | 1.12 |
ENSDART00000166141
|
tekt1
|
tektin 1 |
| chr5_-_23317477 | 1.11 |
ENSDART00000090171
|
nlgn3b
|
neuroligin 3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx3_rfx2+rfx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 1.0 | 2.9 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.9 | 4.7 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.9 | 8.5 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.8 | 2.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.7 | 4.7 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.6 | 3.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.6 | 3.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.5 | 11.1 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.5 | 1.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.4 | 2.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 19.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.4 | 1.5 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.4 | 2.6 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.3 | 1.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 4.7 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.3 | 3.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.3 | 0.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.3 | 1.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.3 | 2.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.3 | 1.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 0.8 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 0.3 | 1.8 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.3 | 1.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.2 | 1.4 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 3.3 | GO:0030317 | sperm motility(GO:0030317) |
| 0.2 | 2.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 3.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 2.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 8.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 1.0 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.2 | 2.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 2.0 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.2 | 2.8 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 1.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 1.6 | GO:1902110 | positive regulation of mitochondrial membrane permeability(GO:0035794) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.2 | 2.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.2 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 1.0 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.7 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 1.7 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 1.1 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 3.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.4 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 8.7 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.1 | 0.9 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.1 | 0.8 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.6 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 5.3 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 2.6 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.8 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 4.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 1.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.6 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 3.1 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 4.6 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 0.3 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 1.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 2.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.6 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.1 | 1.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.2 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 2.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.8 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 1.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 1.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.6 | GO:0050796 | regulation of insulin secretion(GO:0050796) hepatocyte differentiation(GO:0070365) |
| 0.1 | 2.3 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.2 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.1 | 2.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.9 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.8 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.6 | GO:0035094 | response to nicotine(GO:0035094) response to alkaloid(GO:0043279) |
| 0.0 | 1.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.5 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 2.5 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.5 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 3.2 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.0 | 3.5 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 0.4 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.4 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.7 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.4 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 1.9 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 2.9 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 4.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 2.1 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 1.4 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 1.7 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 3.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.2 | GO:1901490 | protein kinase D signaling(GO:0089700) regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 1.2 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 | 1.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 3.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.2 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 7.6 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.4 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 2.2 | GO:0030900 | forebrain development(GO:0030900) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.8 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 1.4 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.2 | GO:0010574 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0015740 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) C4-dicarboxylate transport(GO:0015740) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0001534 | radial spoke(GO:0001534) |
| 0.4 | 1.6 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.4 | 3.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 5.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.3 | 3.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 3.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.3 | 11.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 0.9 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 1.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 2.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 1.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 2.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 3.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 18.2 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 3.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 2.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 3.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 8.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.7 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 0.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 3.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 3.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 2.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 2.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 9.5 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.3 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 3.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 6.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 3.1 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 13.4 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.4 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.1 | 5.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.7 | 2.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.6 | 3.0 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.4 | 5.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 1.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 4.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.3 | 5.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 0.8 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.2 | 2.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 2.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 1.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 2.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 5.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.2 | 2.8 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 6.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 2.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.8 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 2.1 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 3.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 8.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 5.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 3.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 2.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 3.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 8.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.5 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 26.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 2.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 2.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.4 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.1 | GO:0051393 | actinin binding(GO:0042805) muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.5 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 2.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.0 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 1.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 2.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.6 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 6.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.3 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors(GO:0016614) |
| 0.0 | 13.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 22.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 6.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 1.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 3.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 22.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.5 | 2.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 5.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 2.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.4 | 5.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 5.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.3 | 5.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 4.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 11.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 1.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 0.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 0.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 4.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |