Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
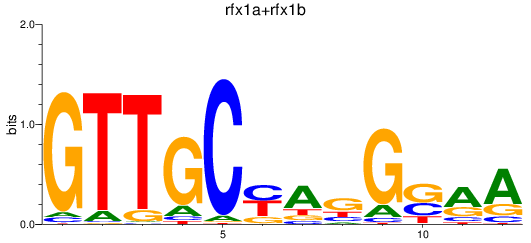
Results for rfx1a+rfx1b
Z-value: 0.37
Transcription factors associated with rfx1a+rfx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rfx1a
|
ENSDARG00000005883 | regulatory factor X, 1a (influences HLA class II expression) |
|
rfx1b
|
ENSDARG00000075904 | regulatory factor X, 1b (influences HLA class II expression) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rfx1a | dr11_v1_chr3_-_19200571_19200651 | -0.69 | 4.1e-02 | Click! |
| rfx1b | dr11_v1_chr1_-_54107321_54107321 | 0.58 | 1.0e-01 | Click! |
Activity profile of rfx1a+rfx1b motif
Sorted Z-values of rfx1a+rfx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_35892767 | 0.56 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr4_-_20177868 | 0.50 |
ENSDART00000003621
|
sinup
|
siaz-interacting nuclear protein |
| chr13_+_48482256 | 0.35 |
ENSDART00000053332
|
FOXN2 (1 of many)
|
forkhead box N2 |
| chr19_-_27588842 | 0.35 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr15_-_47193564 | 0.33 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr22_-_24738188 | 0.31 |
ENSDART00000050238
|
vtg1
|
vitellogenin 1 |
| chr14_-_33481428 | 0.30 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr21_+_3244146 | 0.30 |
ENSDART00000127740
|
ctif
|
CBP80/20-dependent translation initiation factor |
| chr25_+_34407740 | 0.30 |
ENSDART00000012677
|
OTUD7A
|
si:dkey-37f18.2 |
| chr10_-_44560165 | 0.27 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr2_-_16449504 | 0.27 |
ENSDART00000144801
|
atr
|
ATR serine/threonine kinase |
| chr6_+_18418651 | 0.25 |
ENSDART00000171097
ENSDART00000167798 |
rab11fip4b
|
RAB11 family interacting protein 4 (class II) b |
| chr19_+_10661520 | 0.25 |
ENSDART00000091813
ENSDART00000165653 |
ago3b
|
argonaute RISC catalytic component 3b |
| chr14_-_897874 | 0.25 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr13_-_51952020 | 0.24 |
ENSDART00000181079
|
LT631676.1
|
|
| chr1_+_604127 | 0.24 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
| chr12_-_48168135 | 0.23 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr14_+_30413312 | 0.23 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr5_-_72390259 | 0.23 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr1_+_594584 | 0.22 |
ENSDART00000135944
|
jam2a
|
junctional adhesion molecule 2a |
| chr7_-_19999152 | 0.21 |
ENSDART00000173881
ENSDART00000100798 |
trip6
|
thyroid hormone receptor interactor 6 |
| chr13_+_35528607 | 0.21 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr17_+_12075805 | 0.21 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr3_+_17456428 | 0.21 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr6_+_515181 | 0.20 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr24_-_1657276 | 0.20 |
ENSDART00000168131
|
si:ch73-378g22.1
|
si:ch73-378g22.1 |
| chr15_+_31735931 | 0.20 |
ENSDART00000185681
ENSDART00000149137 |
rxfp2b
|
relaxin/insulin-like family peptide receptor 2b |
| chr13_-_24717365 | 0.20 |
ENSDART00000137934
ENSDART00000003922 |
erlin1
|
ER lipid raft associated 1 |
| chr5_-_45958838 | 0.20 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr16_-_41717063 | 0.20 |
ENSDART00000084610
|
cep85
|
centrosomal protein 85 |
| chr3_-_6719232 | 0.20 |
ENSDART00000154294
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr11_-_18799827 | 0.20 |
ENSDART00000185438
ENSDART00000189116 ENSDART00000180504 |
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr8_-_22274222 | 0.19 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr15_-_28587147 | 0.19 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr21_-_32781612 | 0.19 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr9_-_41401564 | 0.19 |
ENSDART00000059628
|
nab1b
|
NGFI-A binding protein 1b (EGR1 binding protein 1) |
| chr6_-_41138854 | 0.19 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr15_-_37589600 | 0.19 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr16_+_11779761 | 0.19 |
ENSDART00000140297
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr5_-_38384755 | 0.18 |
ENSDART00000188573
ENSDART00000051233 |
mink1
|
misshapen-like kinase 1 |
| chr20_-_32007209 | 0.18 |
ENSDART00000021575
|
adgb
|
androglobin |
| chr16_+_11779534 | 0.18 |
ENSDART00000133497
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr13_-_24717618 | 0.17 |
ENSDART00000172156
|
erlin1
|
ER lipid raft associated 1 |
| chr10_+_6907715 | 0.17 |
ENSDART00000041068
|
slc38a9
|
solute carrier family 38, member 9 |
| chr20_+_49787584 | 0.17 |
ENSDART00000193458
ENSDART00000181511 ENSDART00000185850 ENSDART00000185613 ENSDART00000191671 |
CABZ01078261.1
|
|
| chr15_-_28587490 | 0.17 |
ENSDART00000186196
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr22_-_10891213 | 0.17 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr8_-_22274509 | 0.17 |
ENSDART00000181805
ENSDART00000100063 |
nphp4
|
nephronophthisis 4 |
| chr2_+_41526904 | 0.17 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr12_+_46582727 | 0.17 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr17_+_34186632 | 0.16 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr8_-_9570511 | 0.16 |
ENSDART00000044000
|
plxna3
|
plexin A3 |
| chr10_+_15255012 | 0.16 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr2_-_58183499 | 0.16 |
ENSDART00000172281
ENSDART00000186262 |
si:ch1073-185p12.2
|
si:ch1073-185p12.2 |
| chr25_+_18436301 | 0.16 |
ENSDART00000056180
|
cep41
|
centrosomal protein 41 |
| chr5_-_38384289 | 0.15 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr17_+_43595692 | 0.15 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr10_+_15608326 | 0.15 |
ENSDART00000188770
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr18_-_44359726 | 0.15 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr22_-_37565348 | 0.15 |
ENSDART00000149482
ENSDART00000104478 |
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr18_+_50880096 | 0.15 |
ENSDART00000169782
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr13_+_24679674 | 0.15 |
ENSDART00000033090
ENSDART00000139854 |
zgc:66426
|
zgc:66426 |
| chr20_-_2361226 | 0.15 |
ENSDART00000172130
|
EPB41L2
|
si:ch73-18b11.1 |
| chr13_-_18691041 | 0.15 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr7_+_23515966 | 0.14 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr21_+_13387965 | 0.14 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr22_-_37834312 | 0.14 |
ENSDART00000076128
|
ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr11_+_37652870 | 0.14 |
ENSDART00000129918
|
kif17
|
kinesin family member 17 |
| chr3_+_60034194 | 0.14 |
ENSDART00000189858
ENSDART00000153642 |
CR352258.1
|
|
| chr4_+_4079418 | 0.13 |
ENSDART00000028016
|
waslb
|
Wiskott-Aldrich syndrome-like b |
| chr18_-_49283058 | 0.13 |
ENSDART00000076554
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr2_+_1988036 | 0.13 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr10_+_15255198 | 0.13 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr13_-_14926318 | 0.13 |
ENSDART00000142785
|
cdc25b
|
cell division cycle 25B |
| chr22_-_16270462 | 0.13 |
ENSDART00000105681
|
cdc14ab
|
cell division cycle 14Ab |
| chr1_+_53279861 | 0.13 |
ENSDART00000035713
|
rnf150a
|
ring finger protein 150a |
| chr3_-_46410387 | 0.13 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr13_-_48764180 | 0.13 |
ENSDART00000167157
|
si:ch1073-266p11.2
|
si:ch1073-266p11.2 |
| chr22_-_16270071 | 0.13 |
ENSDART00000171823
|
cdc14ab
|
cell division cycle 14Ab |
| chr10_-_17550239 | 0.13 |
ENSDART00000057513
|
ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr24_-_36910224 | 0.12 |
ENSDART00000079233
|
dnaaf3l
|
dynein, axonemal, assembly factor 3 like |
| chr17_+_44756247 | 0.12 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr16_+_41717313 | 0.12 |
ENSDART00000015170
ENSDART00000075937 |
mtdhb
|
metadherin b |
| chr12_-_34852342 | 0.12 |
ENSDART00000152968
|
si:dkey-21c1.1
|
si:dkey-21c1.1 |
| chr9_+_2041535 | 0.12 |
ENSDART00000093187
|
lnpa
|
limb and neural patterns a |
| chr25_-_21894706 | 0.12 |
ENSDART00000189158
|
fbxo31
|
F-box protein 31 |
| chr24_+_39186940 | 0.12 |
ENSDART00000155817
|
spsb3b
|
splA/ryanodine receptor domain and SOCS box containing 3b |
| chr25_+_3868438 | 0.12 |
ENSDART00000156438
|
tmem138
|
transmembrane protein 138 |
| chr15_+_26603395 | 0.12 |
ENSDART00000188667
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr17_+_20569806 | 0.12 |
ENSDART00000113936
|
SAMD8
|
zgc:162183 |
| chr17_+_1496107 | 0.12 |
ENSDART00000187804
|
LO018430.1
|
|
| chr11_-_18800299 | 0.12 |
ENSDART00000156276
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr1_+_30723380 | 0.11 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr2_-_43123949 | 0.11 |
ENSDART00000075347
ENSDART00000132346 |
lrrc6
|
leucine rich repeat containing 6 |
| chr20_+_37825804 | 0.11 |
ENSDART00000152865
|
tatdn3
|
TatD DNase domain containing 3 |
| chr11_+_45092866 | 0.11 |
ENSDART00000163408
|
si:dkey-93h22.8
|
si:dkey-93h22.8 |
| chr2_-_45663945 | 0.11 |
ENSDART00000075080
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr4_-_3145359 | 0.11 |
ENSDART00000112210
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr6_-_7711349 | 0.11 |
ENSDART00000032494
|
ttc26
|
tetratricopeptide repeat domain 26 |
| chr12_+_11650146 | 0.11 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr1_+_30723677 | 0.11 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr4_+_13615731 | 0.11 |
ENSDART00000141395
ENSDART00000144310 |
ccdc87
|
coiled-coil domain containing 87 |
| chr23_+_44665230 | 0.11 |
ENSDART00000144958
|
camta2
|
calmodulin binding transcription activator 2 |
| chr18_-_2727764 | 0.10 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr14_+_35892802 | 0.10 |
ENSDART00000135079
|
zgc:63568
|
zgc:63568 |
| chr16_+_9726038 | 0.10 |
ENSDART00000192988
ENSDART00000020859 |
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr24_+_25822999 | 0.10 |
ENSDART00000109809
|
sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr10_+_2234283 | 0.10 |
ENSDART00000136363
|
cntnap3
|
contactin associated protein like 3 |
| chr3_+_29469283 | 0.10 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| chr21_-_40562705 | 0.10 |
ENSDART00000158289
ENSDART00000171997 |
taok1b
|
TAO kinase 1b |
| chr6_-_58901552 | 0.10 |
ENSDART00000122003
|
mbd6
|
methyl-CpG binding domain protein 6 |
| chr13_+_9368621 | 0.10 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr15_-_43768776 | 0.10 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr24_+_17262879 | 0.10 |
ENSDART00000145949
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr19_+_29337789 | 0.10 |
ENSDART00000021396
|
ldlrap1a
|
low density lipoprotein receptor adaptor protein 1a |
| chr17_+_33296852 | 0.10 |
ENSDART00000154580
|
dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr8_-_22273819 | 0.10 |
ENSDART00000121513
|
nphp4
|
nephronophthisis 4 |
| chr17_-_28811747 | 0.09 |
ENSDART00000001444
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr18_+_5917625 | 0.09 |
ENSDART00000169100
|
glg1b
|
golgi glycoprotein 1b |
| chr17_-_49412313 | 0.09 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr19_+_21266008 | 0.09 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr13_+_13805385 | 0.09 |
ENSDART00000166859
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr3_+_32933663 | 0.09 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr23_+_37185247 | 0.09 |
ENSDART00000146269
|
vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr18_-_43884044 | 0.09 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr21_+_22124736 | 0.09 |
ENSDART00000130179
ENSDART00000172573 |
cul5b
|
cullin 5b |
| chr11_+_36409457 | 0.09 |
ENSDART00000077641
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr12_+_28955766 | 0.09 |
ENSDART00000123417
ENSDART00000139347 |
znf668
|
zinc finger protein 668 |
| chr25_-_3867990 | 0.09 |
ENSDART00000075663
|
cracr2b
|
calcium release activated channel regulator 2B |
| chr2_-_10631767 | 0.09 |
ENSDART00000190033
|
mtf2
|
metal response element binding transcription factor 2 |
| chr20_+_31076488 | 0.09 |
ENSDART00000136255
ENSDART00000008840 |
otofa
|
otoferlin a |
| chr7_-_21905851 | 0.09 |
ENSDART00000111066
ENSDART00000020288 |
epoa
|
erythropoietin a |
| chr19_-_5058908 | 0.09 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr19_+_551963 | 0.09 |
ENSDART00000110495
|
akap9
|
A kinase (PRKA) anchor protein 9 |
| chr6_-_24053404 | 0.08 |
ENSDART00000168511
|
si:dkey-44g17.6
|
si:dkey-44g17.6 |
| chr12_+_33361948 | 0.08 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr17_-_14451405 | 0.08 |
ENSDART00000146786
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr19_-_27395531 | 0.08 |
ENSDART00000103940
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr13_-_4303641 | 0.08 |
ENSDART00000136262
|
ppp2r5d
|
protein phosphatase 2, regulatory subunit B', delta |
| chr6_+_13787855 | 0.08 |
ENSDART00000182899
|
tmem198b
|
transmembrane protein 198b |
| chr2_-_36819624 | 0.08 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr5_+_13394543 | 0.08 |
ENSDART00000051669
ENSDART00000135921 |
tctn2
|
tectonic family member 2 |
| chr5_+_51227147 | 0.08 |
ENSDART00000083340
|
ubac1
|
UBA domain containing 1 |
| chr9_+_25853052 | 0.08 |
ENSDART00000127135
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr18_+_38191346 | 0.08 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr7_+_17907883 | 0.08 |
ENSDART00000147564
|
mta2
|
metastasis associated 1 family, member 2 |
| chr18_+_5454341 | 0.07 |
ENSDART00000192649
|
dtwd1
|
DTW domain containing 1 |
| chr8_-_36327328 | 0.07 |
ENSDART00000183333
|
zgc:103700
|
zgc:103700 |
| chr5_-_69934558 | 0.07 |
ENSDART00000124954
|
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr17_+_37510834 | 0.07 |
ENSDART00000045583
|
mta1
|
metastasis associated 1 |
| chr9_-_40873934 | 0.07 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr20_+_36682051 | 0.07 |
ENSDART00000130513
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr8_-_36469117 | 0.07 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr17_-_14451718 | 0.07 |
ENSDART00000039438
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr13_+_36958086 | 0.07 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr7_+_29163762 | 0.07 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr10_-_76352 | 0.07 |
ENSDART00000186560
ENSDART00000144722 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr23_-_12158685 | 0.07 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr4_-_1801519 | 0.07 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr10_-_74408 | 0.07 |
ENSDART00000100073
ENSDART00000141723 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr17_-_15229787 | 0.07 |
ENSDART00000039165
|
styx
|
serine/threonine/tyrosine interacting protein |
| chr4_-_18606524 | 0.07 |
ENSDART00000173438
ENSDART00000137798 ENSDART00000172976 ENSDART00000171450 ENSDART00000184288 |
CDPF1
|
si:ch211-239e6.4 |
| chr11_+_1575435 | 0.07 |
ENSDART00000155713
ENSDART00000156562 |
si:dkey-40c23.1
|
si:dkey-40c23.1 |
| chr11_+_24800156 | 0.07 |
ENSDART00000131976
|
adipor1a
|
adiponectin receptor 1a |
| chr10_+_2529037 | 0.06 |
ENSDART00000123467
|
zmp:0000001301
|
zmp:0000001301 |
| chr4_+_10616626 | 0.06 |
ENSDART00000067251
ENSDART00000143690 |
cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr5_-_33022014 | 0.06 |
ENSDART00000061149
|
zgc:55461
|
zgc:55461 |
| chr10_-_15854743 | 0.06 |
ENSDART00000092343
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr3_-_36209936 | 0.06 |
ENSDART00000175208
|
csnk1da
|
casein kinase 1, delta a |
| chr20_-_7582936 | 0.06 |
ENSDART00000083890
|
usp24
|
ubiquitin specific peptidase 24 |
| chr2_-_103388 | 0.06 |
ENSDART00000169372
|
angptl1b
|
angiopoietin-like 1b |
| chr7_+_17938128 | 0.06 |
ENSDART00000141044
|
mta2
|
metastasis associated 1 family, member 2 |
| chr8_-_14080534 | 0.06 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr8_+_35356944 | 0.06 |
ENSDART00000143243
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr14_+_7892021 | 0.05 |
ENSDART00000161307
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr25_+_2415131 | 0.05 |
ENSDART00000162210
|
zmp:0000000932
|
zmp:0000000932 |
| chr5_-_66823750 | 0.05 |
ENSDART00000041441
ENSDART00000112488 |
stip1
|
stress-induced phosphoprotein 1 |
| chr25_+_5755314 | 0.05 |
ENSDART00000172088
ENSDART00000164208 |
appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr8_-_22838364 | 0.05 |
ENSDART00000187954
ENSDART00000185368 ENSDART00000188210 |
magixa
|
MAGI family member, X-linked a |
| chr18_-_31051847 | 0.05 |
ENSDART00000170982
|
gas8
|
growth arrest-specific 8 |
| chr15_-_1534232 | 0.05 |
ENSDART00000056763
ENSDART00000133943 |
ift80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr10_-_43721530 | 0.05 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr16_-_31791165 | 0.05 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr13_+_669177 | 0.05 |
ENSDART00000190085
|
CU462915.1
|
|
| chr4_+_20177526 | 0.05 |
ENSDART00000017947
ENSDART00000135451 |
ccdc146
|
coiled-coil domain containing 146 |
| chr17_+_33296436 | 0.05 |
ENSDART00000123114
|
dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr13_+_31205439 | 0.05 |
ENSDART00000132326
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr13_+_24717880 | 0.05 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr19_+_27970292 | 0.05 |
ENSDART00000103887
ENSDART00000145549 |
med10
|
mediator complex subunit 10 |
| chr10_+_25204626 | 0.05 |
ENSDART00000024514
|
chordc1b
|
cysteine and histidine-rich domain (CHORD) containing 1b |
| chr8_+_29749017 | 0.05 |
ENSDART00000185144
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr4_+_74002176 | 0.04 |
ENSDART00000163662
|
si:cabz01069013.3
|
si:cabz01069013.3 |
| chr25_+_186583 | 0.04 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr2_+_307665 | 0.04 |
ENSDART00000082083
|
zgc:113452
|
zgc:113452 |
| chr7_+_21180747 | 0.04 |
ENSDART00000185543
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr1_+_5422439 | 0.04 |
ENSDART00000055047
ENSDART00000142248 |
stk16
|
serine/threonine kinase 16 |
| chr12_+_17620067 | 0.04 |
ENSDART00000073599
|
rsph10b
|
radial spoke head 10 homolog B |
| chr10_-_24753715 | 0.04 |
ENSDART00000192401
|
ilk
|
integrin-linked kinase |
| chr1_-_53277187 | 0.04 |
ENSDART00000131520
|
znf330
|
zinc finger protein 330 |
| chr11_-_28911172 | 0.04 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx1a+rfx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.2 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.3 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.1 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.5 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 0.1 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.4 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.2 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.3 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.0 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.0 | 0.1 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.1 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.2 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.6 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |