Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
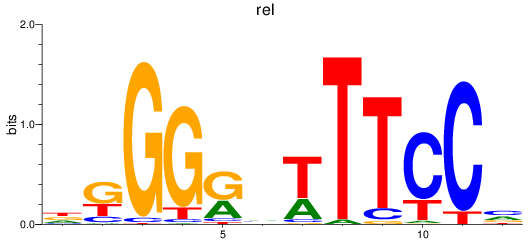
Results for rel
Z-value: 0.52
Transcription factors associated with rel
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rel
|
ENSDARG00000055276 | v-rel avian reticuloendotheliosis viral oncogene homolog |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rel | dr11_v1_chr13_-_25819825_25819825 | 0.75 | 2.0e-02 | Click! |
Activity profile of rel motif
Sorted Z-values of rel motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_16780643 | 1.25 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr2_-_29485408 | 0.81 |
ENSDART00000013411
|
cahz
|
carbonic anhydrase |
| chr22_-_651719 | 0.76 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr13_-_39947335 | 0.70 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr2_+_42191592 | 0.69 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr12_+_27462225 | 0.68 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr2_+_2737422 | 0.68 |
ENSDART00000032459
|
aqp1a.1
|
aquaporin 1a (Colton blood group), tandem duplicate 1 |
| chr12_-_30760971 | 0.61 |
ENSDART00000066257
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr11_+_8129536 | 0.55 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr7_+_53154946 | 0.53 |
ENSDART00000185104
ENSDART00000191559 ENSDART00000160096 |
cdh29
|
cadherin 29 |
| chr24_-_33703504 | 0.52 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr1_+_18716 | 0.50 |
ENSDART00000172454
ENSDART00000161190 |
nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr8_+_47633438 | 0.50 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr8_-_13010011 | 0.49 |
ENSDART00000140969
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr6_-_54815886 | 0.48 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr8_+_22355909 | 0.46 |
ENSDART00000146457
ENSDART00000142883 |
zgc:153631
|
zgc:153631 |
| chr16_+_40575742 | 0.46 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr12_+_2381213 | 0.45 |
ENSDART00000188007
|
LO018238.1
|
|
| chr13_-_22843562 | 0.43 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr22_+_5478353 | 0.43 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr17_-_6382392 | 0.41 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr16_+_40576679 | 0.39 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr3_+_34670076 | 0.38 |
ENSDART00000133457
|
dlx4a
|
distal-less homeobox 4a |
| chr24_+_4373355 | 0.38 |
ENSDART00000179062
ENSDART00000093256 ENSDART00000138943 |
ccny
|
cyclin Y |
| chr7_+_45975537 | 0.38 |
ENSDART00000170253
|
plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr15_-_2803313 | 0.38 |
ENSDART00000060839
|
tgfb1a
|
transforming growth factor, beta 1a |
| chr5_-_53955183 | 0.37 |
ENSDART00000165740
ENSDART00000169284 |
cldn15b
|
claudin 15b |
| chr12_-_34887943 | 0.36 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr13_+_51579851 | 0.36 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr4_-_12795436 | 0.35 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr16_+_32995882 | 0.35 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr10_-_39011514 | 0.35 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr14_+_36885524 | 0.35 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr15_+_27364394 | 0.34 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr5_+_20693724 | 0.34 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr5_-_28679135 | 0.33 |
ENSDART00000193585
|
tnc
|
tenascin C |
| chr13_+_15004398 | 0.32 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| chr16_+_23403602 | 0.32 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr18_-_16924221 | 0.32 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr16_+_13855039 | 0.31 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr22_-_16180467 | 0.31 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr4_-_18954001 | 0.30 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr1_+_12135129 | 0.30 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr3_-_39695856 | 0.30 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr17_+_8175998 | 0.30 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr2_-_30200206 | 0.30 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr5_+_37032038 | 0.30 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr9_-_18716 | 0.29 |
ENSDART00000164763
|
CABZ01078737.1
|
|
| chr4_-_25485404 | 0.28 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr20_+_28434196 | 0.28 |
ENSDART00000034245
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr14_-_30897177 | 0.28 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr6_+_22679610 | 0.28 |
ENSDART00000102701
|
zmp:0000000634
|
zmp:0000000634 |
| chr20_-_44496245 | 0.28 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr19_+_7717449 | 0.27 |
ENSDART00000104719
ENSDART00000146747 |
tuft1b
|
tuftelin 1b |
| chr16_-_29714540 | 0.27 |
ENSDART00000067854
|
tnfaip8l2b
|
tumor necrosis factor, alpha-induced protein 8-like 2b |
| chr14_+_3038473 | 0.27 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr11_-_3552067 | 0.27 |
ENSDART00000163656
|
CAMK2N1
|
si:dkey-33m11.6 |
| chr7_-_58952382 | 0.26 |
ENSDART00000167076
|
mrc1a
|
mannose receptor, C type 1a |
| chr12_+_34732262 | 0.25 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr18_-_18543358 | 0.25 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr3_-_54544612 | 0.25 |
ENSDART00000018044
|
angptl6
|
angiopoietin-like 6 |
| chr14_-_25577094 | 0.25 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr12_+_7445595 | 0.24 |
ENSDART00000103536
ENSDART00000152524 |
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr7_+_1383027 | 0.23 |
ENSDART00000143762
|
khnyn
|
KH and NYN domain containing |
| chr23_+_19701587 | 0.23 |
ENSDART00000104425
|
dnase1l1
|
deoxyribonuclease I-like 1 |
| chr24_-_38816725 | 0.23 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr22_+_6293563 | 0.23 |
ENSDART00000063416
|
rnasel2
|
ribonuclease like 2 |
| chr14_+_20893065 | 0.22 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr22_+_661711 | 0.22 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr25_+_5288665 | 0.22 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr15_-_33640890 | 0.22 |
ENSDART00000167705
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr3_-_49815223 | 0.21 |
ENSDART00000181208
|
gcgra
|
glucagon receptor a |
| chr23_+_3591690 | 0.21 |
ENSDART00000180822
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr18_+_30847237 | 0.21 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr4_-_22311610 | 0.20 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr13_-_2010191 | 0.20 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr1_+_33328857 | 0.19 |
ENSDART00000137151
|
mxra5a
|
matrix-remodelling associated 5a |
| chr4_-_12790886 | 0.19 |
ENSDART00000182535
ENSDART00000067131 ENSDART00000186426 |
irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr13_-_25819825 | 0.19 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr17_+_24613255 | 0.19 |
ENSDART00000064738
|
atp5if1b
|
ATP synthase inhibitory factor subunit 1b |
| chr1_+_33322555 | 0.18 |
ENSDART00000113486
|
mxra5a
|
matrix-remodelling associated 5a |
| chr15_-_8319988 | 0.18 |
ENSDART00000137552
|
si:ch211-113p18.3
|
si:ch211-113p18.3 |
| chr4_-_25485214 | 0.18 |
ENSDART00000159941
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr23_+_3618454 | 0.17 |
ENSDART00000189393
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr24_-_18477672 | 0.17 |
ENSDART00000126928
ENSDART00000158393 |
si:dkey-73n8.3
|
si:dkey-73n8.3 |
| chr25_-_27843066 | 0.17 |
ENSDART00000179684
ENSDART00000186000 ENSDART00000190065 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr13_+_43400443 | 0.17 |
ENSDART00000084321
|
dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr20_-_33976937 | 0.17 |
ENSDART00000136834
|
sele
|
selectin E |
| chr9_-_98982 | 0.16 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr24_+_22731228 | 0.16 |
ENSDART00000146733
|
si:dkey-225k4.1
|
si:dkey-225k4.1 |
| chr16_-_26296477 | 0.16 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr8_+_24281512 | 0.16 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr12_-_26430507 | 0.16 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr3_-_21242460 | 0.16 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr4_-_23839789 | 0.16 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr10_+_2799285 | 0.16 |
ENSDART00000030709
|
pnx
|
posterior neuron-specific homeobox |
| chr15_+_37436430 | 0.16 |
ENSDART00000124779
|
igflr1
|
IGF-like family receptor 1 |
| chr24_-_18467064 | 0.15 |
ENSDART00000081946
|
zgc:112332
|
zgc:112332 |
| chr23_-_38054 | 0.15 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr4_+_36489448 | 0.15 |
ENSDART00000143181
|
znf1149
|
zinc finger protein 1149 |
| chr5_-_10236599 | 0.15 |
ENSDART00000099834
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr25_-_28926631 | 0.15 |
ENSDART00000112850
|
etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr17_+_23255365 | 0.14 |
ENSDART00000180277
|
AL935174.5
|
|
| chr16_+_43152727 | 0.14 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr23_-_37835794 | 0.14 |
ENSDART00000137358
|
cd40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr18_-_16922905 | 0.14 |
ENSDART00000187165
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr3_-_32590164 | 0.14 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr13_+_41819817 | 0.14 |
ENSDART00000185778
|
CABZ01066611.1
|
|
| chr13_-_133520 | 0.14 |
ENSDART00000180874
|
FO904997.1
|
|
| chr18_-_49286381 | 0.14 |
ENSDART00000174248
ENSDART00000174038 |
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr6_+_202367 | 0.14 |
ENSDART00000168232
|
mgat3b
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase b |
| chr5_+_27525477 | 0.13 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr16_+_38394371 | 0.13 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr23_+_5977965 | 0.13 |
ENSDART00000115403
ENSDART00000183147 |
NAV1 (1 of many)
|
neuron navigator 1 |
| chr6_-_436658 | 0.13 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr7_-_57905274 | 0.13 |
ENSDART00000145925
|
ank2b
|
ankyrin 2b, neuronal |
| chr23_+_3620618 | 0.13 |
ENSDART00000190126
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr13_+_27316632 | 0.13 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr17_+_49081828 | 0.13 |
ENSDART00000156492
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr9_-_5263947 | 0.13 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr25_+_8955530 | 0.13 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr20_+_16881883 | 0.13 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr25_+_37397031 | 0.13 |
ENSDART00000193643
ENSDART00000169132 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr3_-_58733718 | 0.13 |
ENSDART00000154603
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr14_-_33218418 | 0.12 |
ENSDART00000163046
|
si:dkey-31j3.11
|
si:dkey-31j3.11 |
| chr11_-_236984 | 0.12 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr1_-_49409549 | 0.12 |
ENSDART00000074317
|
GSK3B (1 of many)
|
si:dkeyp-80c12.7 |
| chr20_-_39367895 | 0.12 |
ENSDART00000136476
ENSDART00000021788 ENSDART00000180784 |
pbk
|
PDZ binding kinase |
| chr24_-_14711597 | 0.12 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr14_+_36889893 | 0.12 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr11_-_40101246 | 0.12 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr17_-_10315644 | 0.12 |
ENSDART00000168572
|
ttc6
|
tetratricopeptide repeat domain 6 |
| chr25_-_13188214 | 0.11 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr24_+_80653 | 0.11 |
ENSDART00000158473
ENSDART00000129135 |
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr23_+_3596401 | 0.11 |
ENSDART00000185908
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr15_+_32821392 | 0.11 |
ENSDART00000158272
|
dclk1b
|
doublecortin-like kinase 1b |
| chr19_+_23298037 | 0.11 |
ENSDART00000183948
|
irgf1
|
immunity-related GTPase family, f1 |
| chr3_+_20404912 | 0.11 |
ENSDART00000104065
|
nxph3
|
neurexophilin 3 |
| chr23_-_24190712 | 0.11 |
ENSDART00000139514
|
ano11
|
anoctamin 11 |
| chr2_-_14798295 | 0.10 |
ENSDART00000143430
ENSDART00000145869 |
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr21_+_5862476 | 0.10 |
ENSDART00000065848
|
wdr38
|
WD repeat domain 38 |
| chr20_-_20410029 | 0.10 |
ENSDART00000192177
ENSDART00000063483 |
prkchb
|
protein kinase C, eta, b |
| chr4_-_76488581 | 0.10 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr11_-_29737088 | 0.10 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr13_-_27916439 | 0.10 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr4_+_57580303 | 0.10 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr24_-_29997145 | 0.10 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr23_+_3594171 | 0.10 |
ENSDART00000159609
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr5_-_30984271 | 0.10 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr20_+_22045089 | 0.10 |
ENSDART00000063564
ENSDART00000187013 ENSDART00000161552 ENSDART00000174478 ENSDART00000063568 ENSDART00000152247 |
nmu
|
neuromedin U |
| chr14_-_11529311 | 0.09 |
ENSDART00000127208
|
si:ch211-153b23.7
|
si:ch211-153b23.7 |
| chr1_+_16396870 | 0.09 |
ENSDART00000189245
ENSDART00000113266 |
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr22_-_8536482 | 0.09 |
ENSDART00000148134
|
si:ch73-27e22.2
|
si:ch73-27e22.2 |
| chr8_+_48603398 | 0.09 |
ENSDART00000074900
|
zgc:195023
|
zgc:195023 |
| chr14_-_30967284 | 0.09 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr13_+_27316934 | 0.09 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr20_+_26947800 | 0.09 |
ENSDART00000062066
|
si:dkey-177p2.6
|
si:dkey-177p2.6 |
| chr11_+_2156430 | 0.09 |
ENSDART00000182896
|
hoxc13b
|
homeobox C13b |
| chr25_-_28926330 | 0.09 |
ENSDART00000155173
|
etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr21_-_2348838 | 0.09 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr22_+_38301365 | 0.09 |
ENSDART00000137339
|
traf5
|
Tnf receptor-associated factor 5 |
| chr12_-_19250854 | 0.09 |
ENSDART00000152844
|
cdc42ep1a
|
CDC42 effector protein (Rho GTPase binding) 1a |
| chr15_+_791220 | 0.09 |
ENSDART00000136137
|
si:dkey-7i4.1
|
si:dkey-7i4.1 |
| chr5_+_25596234 | 0.09 |
ENSDART00000143949
ENSDART00000167193 |
si:dkey-96l17.6
|
si:dkey-96l17.6 |
| chr8_+_18101608 | 0.08 |
ENSDART00000140941
|
glis1b
|
GLIS family zinc finger 1b |
| chr14_+_21007957 | 0.08 |
ENSDART00000162316
|
pimr194
|
Pim proto-oncogene, serine/threonine kinase, related 194 |
| chr1_+_18550864 | 0.08 |
ENSDART00000142515
|
si:dkey-192k22.2
|
si:dkey-192k22.2 |
| chr11_-_7156620 | 0.08 |
ENSDART00000172823
ENSDART00000172879 ENSDART00000078916 |
smim7
|
small integral membrane protein 7 |
| chr3_-_52644737 | 0.08 |
ENSDART00000126180
|
si:dkey-210j14.3
|
si:dkey-210j14.3 |
| chr1_+_17892944 | 0.08 |
ENSDART00000013021
|
tlr3
|
toll-like receptor 3 |
| chr20_+_33981946 | 0.08 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr12_+_46745239 | 0.08 |
ENSDART00000057179
|
plaub
|
plasminogen activator, urokinase b |
| chr6_-_442163 | 0.08 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr3_-_5555500 | 0.08 |
ENSDART00000176133
ENSDART00000057464 |
trim35-31
|
tripartite motif containing 35-31 |
| chr22_-_8499778 | 0.08 |
ENSDART00000142219
|
si:ch73-27e22.4
|
si:ch73-27e22.4 |
| chr1_+_16397063 | 0.07 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr3_+_46459540 | 0.07 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr9_+_993477 | 0.07 |
ENSDART00000182045
|
CABZ01066921.1
|
|
| chr2_+_6963296 | 0.07 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr4_+_77681389 | 0.07 |
ENSDART00000099727
|
gbp4
|
guanylate binding protein 4 |
| chr4_+_12308976 | 0.07 |
ENSDART00000123953
|
tmem121b
|
transmembrane protein 121B |
| chr6_-_10305918 | 0.07 |
ENSDART00000090994
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr18_+_7073130 | 0.07 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr20_-_51917771 | 0.07 |
ENSDART00000147240
|
dusp10
|
dual specificity phosphatase 10 |
| chr8_-_48704050 | 0.06 |
ENSDART00000163916
|
pimr182
|
Pim proto-oncogene, serine/threonine kinase, related 182 |
| chr25_-_35960229 | 0.06 |
ENSDART00000073434
|
snx20
|
sorting nexin 20 |
| chr10_+_22536693 | 0.06 |
ENSDART00000185037
ENSDART00000146040 |
efnb3a
|
ephrin-B3a |
| chr9_-_52999318 | 0.06 |
ENSDART00000176601
|
CABZ01054965.1
|
|
| chr13_-_36034582 | 0.06 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr10_+_40756352 | 0.06 |
ENSDART00000156210
ENSDART00000144576 |
taar19f
|
trace amine associated receptor 19f |
| chr4_-_149334 | 0.06 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr4_+_8016457 | 0.06 |
ENSDART00000014036
|
optn
|
optineurin |
| chr1_-_37377509 | 0.06 |
ENSDART00000113542
|
tnip2
|
TNFAIP3 interacting protein 2 |
| chr4_-_71708855 | 0.05 |
ENSDART00000158819
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr4_+_55059525 | 0.05 |
ENSDART00000128531
|
si:dkey-19c16.12
|
si:dkey-19c16.12 |
| chr6_-_6448519 | 0.05 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr4_+_69888328 | 0.05 |
ENSDART00000170163
|
si:ch211-145h19.5
|
si:ch211-145h19.5 |
| chr13_-_7474324 | 0.05 |
ENSDART00000162550
|
si:dkey-196h17.9
|
si:dkey-196h17.9 |
| chr2_-_51772438 | 0.05 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr22_-_23067859 | 0.05 |
ENSDART00000137873
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
Network of associatons between targets according to the STRING database.
First level regulatory network of rel
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 0.8 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.2 | 0.7 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.2 | 0.2 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.1 | 0.4 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.1 | 0.3 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.1 | 1.2 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.3 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.1 | 0.6 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.7 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 0.3 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.2 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.4 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.7 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.1 | GO:0035666 | response to protozoan(GO:0001562) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) defense response to protozoan(GO:0042832) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.3 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.3 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 0.1 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 0.1 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.0 | 0.5 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.4 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.1 | GO:0006110 | regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) regulation of cofactor metabolic process(GO:0051193) regulation of coenzyme metabolic process(GO:0051196) |
| 0.0 | 0.4 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.4 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.9 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.0 | GO:0007567 | parturition(GO:0007567) maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.0 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0042613 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.7 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.6 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.0 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |