Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
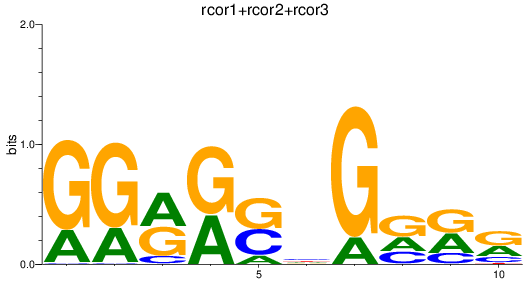
Results for rcor1+rcor2+rcor3
Z-value: 0.41
Transcription factors associated with rcor1+rcor2+rcor3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rcor3
|
ENSDARG00000004502 | REST corepressor 3 |
|
rcor2
|
ENSDARG00000008278 | REST corepressor 2 |
|
rcor1
|
ENSDARG00000031434 | REST corepressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rcor2 | dr11_v1_chr7_-_24994722_24994722 | 0.33 | 3.9e-01 | Click! |
| rcor1 | dr11_v1_chr17_-_29271359_29271359 | 0.25 | 5.1e-01 | Click! |
| rcor3 | dr11_v1_chr22_+_38276024_38276024 | 0.13 | 7.4e-01 | Click! |
Activity profile of rcor1+rcor2+rcor3 motif
Sorted Z-values of rcor1+rcor2+rcor3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_8629275 | 0.44 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr9_-_30371247 | 0.42 |
ENSDART00000079068
|
asb11
|
ankyrin repeat and SOCS box containing 11 |
| chr17_-_8312923 | 0.39 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr2_-_56095275 | 0.29 |
ENSDART00000154701
|
si:ch211-178n15.1
|
si:ch211-178n15.1 |
| chr19_+_14059349 | 0.26 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr10_+_19596214 | 0.25 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr13_-_12021566 | 0.25 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr7_+_5955160 | 0.25 |
ENSDART00000111053
|
zgc:171759
|
zgc:171759 |
| chr17_+_26783828 | 0.24 |
ENSDART00000154921
|
larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr14_+_15484544 | 0.22 |
ENSDART00000188649
|
CR382326.1
|
|
| chr25_+_36311333 | 0.20 |
ENSDART00000190174
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr7_+_39720317 | 0.20 |
ENSDART00000164996
|
si:dkey-58i8.5
|
si:dkey-58i8.5 |
| chr6_-_1820606 | 0.19 |
ENSDART00000183228
|
FO834857.1
|
|
| chr17_-_2584423 | 0.18 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr14_+_14806851 | 0.18 |
ENSDART00000169235
|
fhdc2
|
FH2 domain containing 2 |
| chr22_+_569565 | 0.18 |
ENSDART00000037069
|
usp49
|
ubiquitin specific peptidase 49 |
| chr21_+_21012210 | 0.18 |
ENSDART00000190317
|
ndr1
|
nodal-related 1 |
| chr10_-_35177257 | 0.17 |
ENSDART00000077426
|
pom121
|
POM121 transmembrane nucleoporin |
| chr17_-_2595736 | 0.16 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr12_-_33893381 | 0.16 |
ENSDART00000153280
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr22_-_7050 | 0.16 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr9_-_27717006 | 0.16 |
ENSDART00000146860
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr10_-_7347311 | 0.15 |
ENSDART00000168885
|
nrg1
|
neuregulin 1 |
| chr21_-_11646878 | 0.15 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr9_-_1639884 | 0.15 |
ENSDART00000062850
ENSDART00000150947 |
agps
|
alkylglycerone phosphate synthase |
| chr18_-_17074155 | 0.15 |
ENSDART00000143304
|
tango6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr1_-_49521407 | 0.15 |
ENSDART00000189845
ENSDART00000143474 |
zp3c
|
zona pellucida glycoprotein 3c |
| chr3_-_9738985 | 0.15 |
ENSDART00000160102
|
CR339059.1
|
|
| chr21_+_44581654 | 0.14 |
ENSDART00000187201
ENSDART00000180360 ENSDART00000191543 ENSDART00000180039 ENSDART00000186308 |
TMEM164
|
transmembrane protein 164 |
| chr16_-_42770064 | 0.14 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr8_+_30664077 | 0.14 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr13_+_33688474 | 0.14 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr13_+_40770628 | 0.13 |
ENSDART00000085846
|
nkx1.2la
|
NK1 transcription factor related 2-like,a |
| chr12_-_2800809 | 0.13 |
ENSDART00000152682
ENSDART00000083784 |
ubtd1b
|
ubiquitin domain containing 1b |
| chr2_-_7185460 | 0.13 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr22_+_110158 | 0.13 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr7_-_5070794 | 0.13 |
ENSDART00000097877
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr21_+_3166988 | 0.13 |
ENSDART00000168163
|
hnrnpabb
|
heterogeneous nuclear ribonucleoprotein A/Bb |
| chr25_+_25085349 | 0.12 |
ENSDART00000192166
|
si:ch73-182e20.4
|
si:ch73-182e20.4 |
| chr8_+_25267903 | 0.12 |
ENSDART00000093090
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr16_+_30316177 | 0.12 |
ENSDART00000109505
|
tmem79b
|
transmembrane protein 79b |
| chr6_+_9421279 | 0.12 |
ENSDART00000161036
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr3_+_60957512 | 0.12 |
ENSDART00000044096
|
helz
|
helicase with zinc finger |
| chr1_+_29741843 | 0.12 |
ENSDART00000136066
|
raph1b
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1b |
| chr17_+_43629008 | 0.12 |
ENSDART00000184185
ENSDART00000181681 |
znf365
|
zinc finger protein 365 |
| chr21_-_233282 | 0.12 |
ENSDART00000157684
|
bxdc2
|
brix domain containing 2 |
| chr7_-_13906409 | 0.12 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr2_+_30032303 | 0.12 |
ENSDART00000151841
|
rbm33b
|
RNA binding motif protein 33b |
| chr22_+_25582707 | 0.12 |
ENSDART00000184440
|
aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr11_-_44999858 | 0.12 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr17_-_50020672 | 0.12 |
ENSDART00000188942
|
filip1a
|
filamin A interacting protein 1a |
| chr5_+_22098591 | 0.12 |
ENSDART00000143676
|
zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr2_-_30987538 | 0.11 |
ENSDART00000148157
|
emilin2a
|
elastin microfibril interfacer 2a |
| chr1_+_14454663 | 0.11 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr11_-_42315748 | 0.11 |
ENSDART00000169853
|
slmapa
|
sarcolemma associated protein a |
| chr22_-_38274188 | 0.11 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr18_+_14307059 | 0.11 |
ENSDART00000186558
|
zgc:173742
|
zgc:173742 |
| chr1_-_45584407 | 0.11 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr16_-_10834756 | 0.11 |
ENSDART00000188503
|
rabac1
|
Rab acceptor 1 (prenylated) |
| chr3_-_38777553 | 0.11 |
ENSDART00000193045
|
znf281a
|
zinc finger protein 281a |
| chr4_+_63928858 | 0.11 |
ENSDART00000161958
|
si:dkey-179k24.2
|
si:dkey-179k24.2 |
| chr25_-_35110695 | 0.11 |
ENSDART00000099859
|
hist1h2a10
|
histone cluster 1 H2A family member 10 |
| chr14_+_32964 | 0.11 |
ENSDART00000166173
|
lyar
|
Ly1 antibody reactive homolog (mouse) |
| chr9_+_13628635 | 0.11 |
ENSDART00000108844
ENSDART00000128630 |
als2a
|
amyotrophic lateral sclerosis 2a (juvenile) |
| chr23_+_44236855 | 0.11 |
ENSDART00000130147
ENSDART00000051907 |
MEPCE
|
si:ch1073-157b13.1 |
| chr20_-_32981053 | 0.11 |
ENSDART00000138708
|
nbas
|
neuroblastoma amplified sequence |
| chr22_-_17671348 | 0.11 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr25_-_37258653 | 0.10 |
ENSDART00000131076
|
rfwd3
|
ring finger and WD repeat domain 3 |
| chr13_-_319996 | 0.10 |
ENSDART00000148675
|
ddx21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr15_-_15230264 | 0.10 |
ENSDART00000155400
|
rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr18_+_13077800 | 0.10 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr3_-_27065477 | 0.10 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr19_+_9050852 | 0.10 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr10_+_16168514 | 0.10 |
ENSDART00000175513
|
prrc1
|
proline-rich coiled-coil 1 |
| chr4_+_71401321 | 0.10 |
ENSDART00000158076
|
si:ch211-76m11.7
|
si:ch211-76m11.7 |
| chr17_+_28624321 | 0.10 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr15_+_35933094 | 0.10 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr22_-_26524596 | 0.10 |
ENSDART00000087623
|
zgc:194330
|
zgc:194330 |
| chr8_+_26868105 | 0.10 |
ENSDART00000005337
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr23_+_2717950 | 0.10 |
ENSDART00000137641
|
ncoa6
|
nuclear receptor coactivator 6 |
| chr15_-_25198107 | 0.10 |
ENSDART00000159793
|
mnta
|
MAX network transcriptional repressor a |
| chr4_+_77993977 | 0.10 |
ENSDART00000174118
|
terfa
|
telomeric repeat binding factor a |
| chr18_-_17087138 | 0.10 |
ENSDART00000135597
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr20_-_33556983 | 0.10 |
ENSDART00000168798
|
rock2bl
|
rho-associated, coiled-coil containing protein kinase 2b, like |
| chr12_+_18744610 | 0.10 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr20_-_33675676 | 0.10 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr7_-_56831621 | 0.10 |
ENSDART00000182912
|
senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr17_-_6613458 | 0.10 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr14_+_52408619 | 0.10 |
ENSDART00000163856
|
noa1
|
nitric oxide associated 1 |
| chr10_+_21444654 | 0.09 |
ENSDART00000140113
ENSDART00000184386 ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr25_-_29463866 | 0.09 |
ENSDART00000155676
ENSDART00000124645 ENSDART00000181055 ENSDART00000192593 |
ube2q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr24_+_24170914 | 0.09 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr4_-_890220 | 0.09 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr13_-_31346392 | 0.09 |
ENSDART00000134343
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr13_+_15941850 | 0.09 |
ENSDART00000016294
|
fignl1
|
fidgetin-like 1 |
| chr23_-_6722101 | 0.09 |
ENSDART00000157828
|
baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr25_+_336503 | 0.09 |
ENSDART00000160395
|
CU929262.1
|
|
| chr24_-_25166416 | 0.09 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr4_-_64703 | 0.09 |
ENSDART00000167851
|
CU856344.1
|
|
| chr8_+_48965767 | 0.09 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr21_+_3778498 | 0.09 |
ENSDART00000139813
|
tor1
|
torsin family 1 |
| chr12_-_31457301 | 0.09 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr22_-_10539180 | 0.09 |
ENSDART00000131217
|
ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr20_+_25626479 | 0.09 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr7_+_51795667 | 0.09 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr7_+_21752168 | 0.09 |
ENSDART00000173641
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr3_-_56933578 | 0.09 |
ENSDART00000192185
|
hid1a
|
HID1 domain containing a |
| chr5_-_2672492 | 0.09 |
ENSDART00000192337
|
CABZ01072548.1
|
|
| chr7_-_29223614 | 0.09 |
ENSDART00000173598
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr19_+_14352332 | 0.09 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr22_+_10215558 | 0.09 |
ENSDART00000063274
|
kctd6a
|
potassium channel tetramerization domain containing 6a |
| chr7_-_9674073 | 0.09 |
ENSDART00000187902
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr5_-_31772559 | 0.08 |
ENSDART00000183879
|
fam102ab
|
family with sequence similarity 102, member Ab |
| chr3_+_28502419 | 0.08 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr9_-_54344405 | 0.08 |
ENSDART00000182939
|
CT998556.1
|
|
| chr3_-_15667713 | 0.08 |
ENSDART00000026658
|
zgc:66474
|
zgc:66474 |
| chr10_-_42237304 | 0.08 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr11_+_31324335 | 0.08 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr5_+_24087035 | 0.08 |
ENSDART00000183644
|
tp53
|
tumor protein p53 |
| chr17_-_25630822 | 0.08 |
ENSDART00000126201
ENSDART00000105503 ENSDART00000151878 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr2_-_32826108 | 0.08 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr15_-_434503 | 0.08 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr4_-_965267 | 0.08 |
ENSDART00000093289
|
atp23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr13_-_4367083 | 0.08 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr24_+_18689980 | 0.08 |
ENSDART00000160941
ENSDART00000180303 ENSDART00000190435 |
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr13_-_18119696 | 0.08 |
ENSDART00000148125
|
washc2c
|
WASH complex subunit 2C |
| chr25_+_2993855 | 0.08 |
ENSDART00000163009
|
neo1b
|
neogenin 1b |
| chr19_+_4892281 | 0.08 |
ENSDART00000150969
|
cdk12
|
cyclin-dependent kinase 12 |
| chr8_-_44611357 | 0.08 |
ENSDART00000063396
|
bag4
|
BCL2 associated athanogene 4 |
| chr18_+_17754510 | 0.08 |
ENSDART00000189372
ENSDART00000135236 ENSDART00000137239 |
si:ch211-216l23.2
|
si:ch211-216l23.2 |
| chr14_+_16287968 | 0.08 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr15_+_9327252 | 0.08 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr3_+_37827373 | 0.08 |
ENSDART00000039517
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr25_+_236753 | 0.08 |
ENSDART00000153663
|
pus7
|
pseudouridylate synthase 7 (putative) |
| chr4_-_4250317 | 0.08 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr1_+_36552 | 0.07 |
ENSDART00000169685
|
hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr19_-_31765615 | 0.07 |
ENSDART00000103636
|
si:dkeyp-120h9.1
|
si:dkeyp-120h9.1 |
| chr11_-_44030962 | 0.07 |
ENSDART00000171910
|
FP016005.1
|
|
| chr12_-_4475890 | 0.07 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr5_+_32009956 | 0.07 |
ENSDART00000188482
|
scai
|
suppressor of cancer cell invasion |
| chr5_-_8907819 | 0.07 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr5_-_32505276 | 0.07 |
ENSDART00000034705
ENSDART00000187597 |
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr25_+_236580 | 0.07 |
ENSDART00000040499
|
pus7
|
pseudouridylate synthase 7 (putative) |
| chr8_-_36554675 | 0.07 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr21_-_41369539 | 0.07 |
ENSDART00000187546
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr19_-_11315224 | 0.07 |
ENSDART00000104933
|
eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr25_-_21031007 | 0.07 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr21_+_39941559 | 0.07 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr1_-_40102836 | 0.07 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr2_+_50094873 | 0.07 |
ENSDART00000132307
|
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr3_-_24205339 | 0.07 |
ENSDART00000157135
|
si:dkey-110g7.8
|
si:dkey-110g7.8 |
| chr18_+_11970987 | 0.07 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr2_+_23006792 | 0.07 |
ENSDART00000027782
|
mknk2a
|
MAP kinase interacting serine/threonine kinase 2a |
| chr21_-_15200556 | 0.07 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr8_+_11687254 | 0.07 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr12_+_18971100 | 0.07 |
ENSDART00000164212
|
l3mbtl2
|
l(3)mbt-like 2 (Drosophila) |
| chr5_-_32505109 | 0.07 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr7_+_13491452 | 0.07 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr22_+_22438783 | 0.07 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr13_+_33368503 | 0.07 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr2_+_1989941 | 0.07 |
ENSDART00000190814
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr3_+_15776446 | 0.07 |
ENSDART00000146651
|
znf652
|
zinc finger protein 652 |
| chr7_-_25132592 | 0.07 |
ENSDART00000192214
|
badb
|
BCL2 associated agonist of cell death b |
| chr21_+_233271 | 0.07 |
ENSDART00000171440
|
dtwd2
|
DTW domain containing 2 |
| chr1_+_45056371 | 0.07 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr6_+_52918537 | 0.07 |
ENSDART00000174229
|
or137-1
|
odorant receptor, family H, subfamily 137, member 1 |
| chr7_-_8315179 | 0.06 |
ENSDART00000184049
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr2_+_45374163 | 0.06 |
ENSDART00000193650
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr14_+_81919 | 0.06 |
ENSDART00000171444
|
stag3
|
stromal antigen 3 |
| chr24_-_25166720 | 0.06 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr17_-_10059557 | 0.06 |
ENSDART00000092209
ENSDART00000161243 |
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr13_+_13033837 | 0.06 |
ENSDART00000079558
|
letm1
|
leucine zipper-EF-hand containing transmembrane protein 1 |
| chr1_+_31657842 | 0.06 |
ENSDART00000057880
|
poll
|
polymerase (DNA directed), lambda |
| chr16_-_19890303 | 0.06 |
ENSDART00000147161
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr13_-_18105278 | 0.06 |
ENSDART00000090475
|
washc2c
|
WASH complex subunit 2C |
| chr15_-_43768776 | 0.06 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr6_+_52895008 | 0.06 |
ENSDART00000136143
|
or137-7
|
odorant receptor, family H, subfamily 137, member 7 |
| chr11_+_11271959 | 0.06 |
ENSDART00000190678
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr5_+_3891485 | 0.06 |
ENSDART00000129329
ENSDART00000091711 |
rpain
|
RPA interacting protein |
| chr17_-_38887424 | 0.06 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr4_-_13255700 | 0.06 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr10_-_641609 | 0.06 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr2_-_22492289 | 0.06 |
ENSDART00000168022
|
gtf2b
|
general transcription factor IIB |
| chr2_+_32826235 | 0.06 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr12_+_19348538 | 0.06 |
ENSDART00000066388
|
tomm22
|
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr23_+_43770149 | 0.06 |
ENSDART00000024313
|
rnf150b
|
ring finger protein 150b |
| chr10_-_26179805 | 0.06 |
ENSDART00000174797
|
trim3b
|
tripartite motif containing 3b |
| chr7_-_33829824 | 0.06 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr3_-_34547000 | 0.06 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr8_+_24861264 | 0.06 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr14_+_31865099 | 0.06 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr17_+_389218 | 0.06 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr20_-_54435287 | 0.06 |
ENSDART00000148632
|
yy1b
|
YY1 transcription factor b |
| chr11_-_25734417 | 0.06 |
ENSDART00000103570
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr5_+_6670945 | 0.06 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr6_+_31684 | 0.06 |
ENSDART00000188853
ENSDART00000184553 |
CZQB01141835.2
|
|
| chr19_+_48060464 | 0.06 |
ENSDART00000123163
|
zgc:85936
|
zgc:85936 |
| chr24_+_30521619 | 0.06 |
ENSDART00000162903
|
dpyda.3
|
dihydropyrimidine dehydrogenase a, tandem duplicate 3 |
| chr11_+_45343245 | 0.05 |
ENSDART00000160904
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr7_+_1467863 | 0.05 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rcor1+rcor2+rcor3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.1 | 0.3 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.2 | GO:0060945 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.1 | 0.2 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:2000191 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.5 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.1 | GO:1901216 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.1 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.0 | 0.1 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.4 | GO:0003128 | heart field specification(GO:0003128) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0033238 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.0 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.0 | 0.0 | GO:0071043 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0019860 | uracil catabolic process(GO:0006212) beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) uracil metabolic process(GO:0019860) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.1 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.0 | 0.1 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |