Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
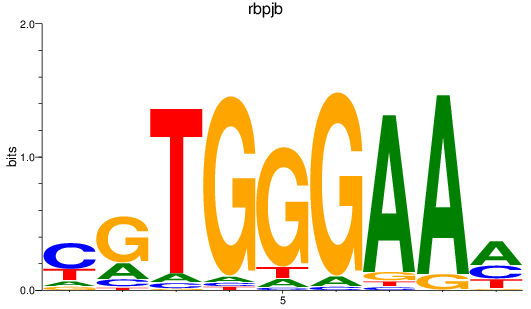
Results for rbpjb
Z-value: 2.37
Transcription factors associated with rbpjb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rbpjb
|
ENSDARG00000052091 | recombination signal binding protein for immunoglobulin kappa J region b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rbpjb | dr11_v1_chr7_+_62080456_62080456 | -0.81 | 7.7e-03 | Click! |
Activity profile of rbpjb motif
Sorted Z-values of rbpjb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_41981959 | 6.77 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr1_+_44442140 | 5.56 |
ENSDART00000190592
|
crybb1l2
|
crystallin, beta B1, like 2 |
| chr7_+_31891110 | 5.36 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr19_-_31802296 | 5.26 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr9_-_22092089 | 5.07 |
ENSDART00000101928
|
crygm2d13
|
crystallin, gamma M2d13 |
| chr24_-_37877743 | 4.89 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr9_-_22205682 | 4.37 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr17_-_25326685 | 4.36 |
ENSDART00000082327
|
pabpc4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr19_+_42609132 | 4.18 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr23_-_23401305 | 4.03 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr3_+_26081343 | 4.01 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr14_+_38786298 | 3.92 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr17_-_25326296 | 3.89 |
ENSDART00000168822
|
pabpc4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr3_+_23677351 | 3.83 |
ENSDART00000023674
|
hoxb9a
|
homeobox B9a |
| chr9_-_22147567 | 3.78 |
ENSDART00000110941
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr19_-_5254699 | 3.62 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr23_-_31372639 | 3.54 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr24_-_26310854 | 3.54 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr25_-_23526058 | 3.53 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr1_-_45633955 | 3.49 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr23_-_21471022 | 3.21 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr9_-_22076368 | 3.19 |
ENSDART00000128486
|
crygm2a
|
crystallin, gamma M2a |
| chr15_+_7176182 | 3.17 |
ENSDART00000101578
|
her8.2
|
hairy-related 8.2 |
| chr19_+_19767567 | 3.10 |
ENSDART00000169074
|
hoxa3a
|
homeobox A3a |
| chr16_-_31717851 | 2.98 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr4_-_20043484 | 2.89 |
ENSDART00000167780
|
zgc:193726
|
zgc:193726 |
| chr5_-_28679135 | 2.88 |
ENSDART00000193585
|
tnc
|
tenascin C |
| chr2_-_32769759 | 2.86 |
ENSDART00000178951
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr7_-_26436436 | 2.83 |
ENSDART00000019035
ENSDART00000123395 |
her8a
|
hairy-related 8a |
| chr13_+_30804367 | 2.81 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr9_-_22345500 | 2.67 |
ENSDART00000101802
|
crygm2f
|
crystallin, gamma M2f |
| chr6_+_3680651 | 2.64 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| chr3_-_36839115 | 2.55 |
ENSDART00000154553
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr7_+_36041509 | 2.49 |
ENSDART00000162850
|
irx3a
|
iroquois homeobox 3a |
| chr22_+_24715282 | 2.48 |
ENSDART00000088027
ENSDART00000189054 ENSDART00000140430 |
ssx2ipb
|
synovial sarcoma, X breakpoint 2 interacting protein b |
| chr16_+_9762261 | 2.46 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr16_-_31718013 | 2.45 |
ENSDART00000190716
|
rbp5
|
retinol binding protein 1a, cellular |
| chr1_+_12763920 | 2.43 |
ENSDART00000189465
|
pcdh10a
|
protocadherin 10a |
| chr9_-_22023061 | 2.41 |
ENSDART00000101952
|
crygm2c
|
crystallin, gamma M2c |
| chr23_-_21446985 | 2.31 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr8_-_13010011 | 2.30 |
ENSDART00000140969
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr5_-_56513825 | 2.23 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr6_+_45932276 | 2.23 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr7_+_46252993 | 2.19 |
ENSDART00000167149
|
znf536
|
zinc finger protein 536 |
| chr24_-_37877554 | 2.10 |
ENSDART00000132219
|
tmem204
|
transmembrane protein 204 |
| chr4_-_9722568 | 2.10 |
ENSDART00000067190
|
tspan9b
|
tetraspanin 9b |
| chr4_-_5302162 | 2.08 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr5_+_10015002 | 2.08 |
ENSDART00000140570
|
slc2a11b
|
solute carrier family 2 (facilitated glucose transporter), member 11b |
| chr5_-_66028714 | 2.07 |
ENSDART00000022625
ENSDART00000164228 |
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr5_+_9348284 | 2.04 |
ENSDART00000149417
|
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr16_+_26449615 | 2.04 |
ENSDART00000039746
|
epb41b
|
erythrocyte membrane protein band 4.1b |
| chr11_-_11518469 | 2.04 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr17_-_6730247 | 1.99 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr23_+_31405497 | 1.99 |
ENSDART00000053546
|
sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr10_+_22782522 | 1.95 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr2_+_35728033 | 1.95 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr2_-_9646857 | 1.93 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr5_+_39563301 | 1.92 |
ENSDART00000131245
ENSDART00000097872 |
si:ch211-57h10.1
|
si:ch211-57h10.1 |
| chr14_+_4151379 | 1.91 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr4_+_21741228 | 1.90 |
ENSDART00000112035
ENSDART00000127664 |
myf5
|
myogenic factor 5 |
| chr15_-_37846047 | 1.90 |
ENSDART00000184837
|
hsc70
|
heat shock cognate 70 |
| chr4_-_9891874 | 1.90 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr2_+_2110271 | 1.88 |
ENSDART00000148460
|
dspa
|
desmoplakin a |
| chr20_+_6590220 | 1.85 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr19_+_44009473 | 1.84 |
ENSDART00000151004
|
nkd3
|
naked cuticle homolog 3 |
| chr22_+_5727753 | 1.84 |
ENSDART00000063484
|
si:dkey-222f2.1
|
si:dkey-222f2.1 |
| chr14_+_35806605 | 1.79 |
ENSDART00000173093
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr10_+_25982212 | 1.79 |
ENSDART00000128292
ENSDART00000108808 |
frem2a
|
Fras1 related extracellular matrix protein 2a |
| chr5_-_47727819 | 1.75 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr6_-_14139503 | 1.71 |
ENSDART00000089577
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr5_-_33287691 | 1.68 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr13_-_36579086 | 1.66 |
ENSDART00000146671
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr7_+_38445938 | 1.64 |
ENSDART00000173467
|
cep89
|
centrosomal protein 89 |
| chr3_+_23092762 | 1.62 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr3_+_37574885 | 1.62 |
ENSDART00000055225
|
wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr5_+_15202495 | 1.52 |
ENSDART00000144915
|
tbx1
|
T-box 1 |
| chr9_+_38684871 | 1.45 |
ENSDART00000147938
ENSDART00000004462 |
heg1
|
heart development protein with EGF-like domains 1 |
| chr4_+_19535946 | 1.43 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr20_-_47731768 | 1.41 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr13_-_27916439 | 1.40 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr3_+_41917499 | 1.39 |
ENSDART00000028673
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr2_+_3696038 | 1.37 |
ENSDART00000150499
ENSDART00000136906 ENSDART00000164152 ENSDART00000128514 ENSDART00000108964 |
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr8_+_23009662 | 1.35 |
ENSDART00000030885
|
uckl1a
|
uridine-cytidine kinase 1-like 1a |
| chr3_-_29869120 | 1.34 |
ENSDART00000138327
|
rpl3
|
ribosomal protein L3 |
| chr3_-_28665291 | 1.32 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr21_-_5881344 | 1.32 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr4_-_8014463 | 1.32 |
ENSDART00000036153
|
ccdc3a
|
coiled-coil domain containing 3a |
| chr13_+_33246150 | 1.30 |
ENSDART00000136356
ENSDART00000132834 ENSDART00000144379 ENSDART00000131985 ENSDART00000148368 ENSDART00000035940 ENSDART00000141912 |
ndufaf1
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr5_-_2112030 | 1.29 |
ENSDART00000091932
|
gusb
|
glucuronidase, beta |
| chr21_-_5799122 | 1.27 |
ENSDART00000129351
ENSDART00000151202 |
ccni
|
cyclin I |
| chr5_-_66028371 | 1.27 |
ENSDART00000183012
|
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr22_-_11729350 | 1.24 |
ENSDART00000105813
|
krt222
|
keratin 222 |
| chr7_-_33960170 | 1.23 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr23_-_31346319 | 1.22 |
ENSDART00000008401
ENSDART00000138106 |
phip
|
pleckstrin homology domain interacting protein |
| chr7_+_39634873 | 1.22 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr16_+_36671064 | 1.22 |
ENSDART00000109703
|
col6a4a
|
collagen, type VI, alpha 4a |
| chr21_-_43485351 | 1.19 |
ENSDART00000151597
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr16_-_27628994 | 1.17 |
ENSDART00000157407
|
nacad
|
NAC alpha domain containing |
| chr10_+_17026870 | 1.14 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr17_+_24851951 | 1.12 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr19_-_32641725 | 1.11 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr16_+_46492994 | 1.10 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr16_+_5579744 | 1.10 |
ENSDART00000147973
|
macf1b
|
microtubule-actin crosslinking factor 1 b |
| chr2_+_32835679 | 1.08 |
ENSDART00000132767
|
pxdc1a
|
PX domain containing 1a |
| chr22_-_30881738 | 1.08 |
ENSDART00000059965
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr7_+_27834130 | 1.08 |
ENSDART00000052656
|
rras2
|
RAS related 2 |
| chr19_-_7540821 | 1.07 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr9_-_52962521 | 1.06 |
ENSDART00000170419
|
CU855885.1
|
|
| chr19_+_48018464 | 1.05 |
ENSDART00000172307
ENSDART00000163848 |
UBE2M
|
si:ch1073-205c8.3 |
| chr11_-_7320211 | 1.04 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr5_+_46424437 | 1.04 |
ENSDART00000186511
|
vcana
|
versican a |
| chr10_-_10864331 | 1.04 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr3_+_59051503 | 1.04 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr25_+_6186823 | 1.03 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr4_-_25215968 | 1.03 |
ENSDART00000066932
ENSDART00000066933 |
itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr6_-_36552844 | 1.02 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr13_-_40398556 | 1.00 |
ENSDART00000191921
|
nkx3.3
|
NK3 homeobox 3 |
| chr14_+_41887070 | 1.00 |
ENSDART00000173264
|
slc7a11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr10_-_10863936 | 0.99 |
ENSDART00000180568
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr25_+_10793478 | 0.97 |
ENSDART00000058339
ENSDART00000134923 |
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr11_+_13159988 | 0.97 |
ENSDART00000064176
|
mob3c
|
MOB kinase activator 3C |
| chr5_-_37252111 | 0.96 |
ENSDART00000185110
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr8_+_48491387 | 0.95 |
ENSDART00000086053
|
PRDM16
|
si:ch211-263k4.2 |
| chr20_+_39283849 | 0.94 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr16_-_42523744 | 0.94 |
ENSDART00000017185
|
tbx20
|
T-box 20 |
| chr5_-_22082918 | 0.92 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr17_+_6956696 | 0.90 |
ENSDART00000171368
|
zgc:172341
|
zgc:172341 |
| chr25_+_3525494 | 0.86 |
ENSDART00000169775
|
RAB19 (1 of many)
|
RAB19, member RAS oncogene family |
| chr22_+_12431608 | 0.85 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr20_-_26536581 | 0.85 |
ENSDART00000181810
|
stx11b.2
|
syntaxin 11b, tandem duplicate 2 |
| chr8_-_41273768 | 0.83 |
ENSDART00000108518
|
rnf10
|
ring finger protein 10 |
| chr9_-_3671911 | 0.82 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr7_-_40738774 | 0.80 |
ENSDART00000084179
|
rnf32
|
ring finger protein 32 |
| chr19_+_48018802 | 0.80 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr8_+_48848200 | 0.79 |
ENSDART00000130673
|
tp73
|
tumor protein p73 |
| chr8_-_41273461 | 0.78 |
ENSDART00000190598
|
rnf10
|
ring finger protein 10 |
| chr7_+_17229282 | 0.77 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr25_+_21178033 | 0.77 |
ENSDART00000041169
|
erc1a
|
ELKS/RAB6-interacting/CAST family member 1a |
| chr15_+_5321612 | 0.76 |
ENSDART00000174345
|
or113-1
|
odorant receptor, family A, subfamily 113, member 1 |
| chr10_-_4375190 | 0.76 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr10_+_1668106 | 0.76 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr14_+_40685469 | 0.75 |
ENSDART00000172839
|
tnmd
|
tenomodulin |
| chr1_-_51720633 | 0.75 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr11_-_29563437 | 0.73 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr9_-_7089303 | 0.72 |
ENSDART00000146609
|
coa5
|
cytochrome C oxidase assembly factor 5 |
| chr1_-_56363108 | 0.71 |
ENSDART00000021878
|
gcgrb
|
glucagon receptor b |
| chr8_-_15150131 | 0.70 |
ENSDART00000138253
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr17_-_20287530 | 0.66 |
ENSDART00000078703
ENSDART00000191289 |
add3b
|
adducin 3 (gamma) b |
| chr18_-_6943577 | 0.63 |
ENSDART00000132399
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr24_+_33622769 | 0.63 |
ENSDART00000079342
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr25_+_10830269 | 0.63 |
ENSDART00000175736
|
si:ch211-147g22.5
|
si:ch211-147g22.5 |
| chr24_-_6375774 | 0.62 |
ENSDART00000013294
|
anxa13
|
annexin A13 |
| chr9_+_7358749 | 0.61 |
ENSDART00000081660
|
ihha
|
Indian hedgehog homolog a |
| chr23_+_32011768 | 0.61 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr25_+_35304903 | 0.60 |
ENSDART00000034313
ENSDART00000187659 |
gas2a
|
growth arrest-specific 2a |
| chr16_-_27640995 | 0.59 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr14_-_2213660 | 0.58 |
ENSDART00000162537
|
pcdh2ab3
|
protocadherin 2 alpha b 3 |
| chr3_-_27784457 | 0.58 |
ENSDART00000019098
|
dnase1
|
deoxyribonuclease I |
| chr3_+_27722355 | 0.56 |
ENSDART00000132761
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr5_-_57361594 | 0.56 |
ENSDART00000112793
|
cdc26
|
cell division cycle 26 homolog |
| chr16_-_17300030 | 0.56 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr9_-_56399699 | 0.55 |
ENSDART00000170281
|
rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
| chr2_-_44746723 | 0.55 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium chain family member 3 |
| chr16_-_40727455 | 0.55 |
ENSDART00000162331
|
si:dkey-22o22.2
|
si:dkey-22o22.2 |
| chr5_+_9360394 | 0.54 |
ENSDART00000124642
|
FP236810.2
|
|
| chr15_+_5339056 | 0.53 |
ENSDART00000174319
|
CABZ01018874.2
|
|
| chr7_+_19424857 | 0.51 |
ENSDART00000173674
|
si:ch211-212k18.6
|
si:ch211-212k18.6 |
| chr7_-_71837213 | 0.50 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr14_-_33521071 | 0.50 |
ENSDART00000052789
|
c1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr5_+_9377005 | 0.49 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr1_-_38538284 | 0.49 |
ENSDART00000009777
|
glra3
|
glycine receptor, alpha 3 |
| chr10_+_3507861 | 0.48 |
ENSDART00000092684
|
rph3aa
|
rabphilin 3A homolog (mouse), a |
| chr23_-_36003441 | 0.48 |
ENSDART00000164699
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr22_-_26834043 | 0.48 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr18_+_12655766 | 0.46 |
ENSDART00000144246
|
tbxas1
|
thromboxane A synthase 1 (platelet) |
| chr21_+_20549395 | 0.46 |
ENSDART00000181633
|
efna5a
|
ephrin-A5a |
| chr1_+_41886830 | 0.45 |
ENSDART00000185887
ENSDART00000084735 |
si:dkey-37g12.1
|
si:dkey-37g12.1 |
| chr23_+_26039524 | 0.45 |
ENSDART00000142851
|
itih6
|
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr20_-_15090862 | 0.44 |
ENSDART00000063892
ENSDART00000122592 |
si:dkey-239i20.2
|
si:dkey-239i20.2 |
| chr5_-_25072607 | 0.44 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr1_+_32528097 | 0.44 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr9_+_29589790 | 0.43 |
ENSDART00000140388
|
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr13_-_33654931 | 0.43 |
ENSDART00000020350
|
snx5
|
sorting nexin 5 |
| chr5_+_9382301 | 0.41 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr9_-_22355391 | 0.40 |
ENSDART00000009115
|
crygm3
|
crystallin, gamma M3 |
| chr5_+_38837429 | 0.39 |
ENSDART00000160236
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr18_-_21177674 | 0.39 |
ENSDART00000060175
|
si:dkey-12e7.4
|
si:dkey-12e7.4 |
| chr17_+_35362851 | 0.39 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr2_+_6734108 | 0.38 |
ENSDART00000112227
|
brinp3b
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3b |
| chr22_+_30335936 | 0.38 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr10_+_9195190 | 0.37 |
ENSDART00000136364
|
antxr2b
|
anthrax toxin receptor 2b |
| chr15_-_21702317 | 0.36 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr15_+_29362714 | 0.34 |
ENSDART00000099916
ENSDART00000126196 |
gucy2f
|
guanylate cyclase 2F, retinal |
| chr17_+_49081828 | 0.34 |
ENSDART00000156492
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr14_+_30762131 | 0.32 |
ENSDART00000145039
|
si:ch211-145o7.3
|
si:ch211-145o7.3 |
| chr12_+_41991635 | 0.31 |
ENSDART00000186161
ENSDART00000192510 |
TCERG1L
|
transcription elongation regulator 1 like |
| chr6_+_72040 | 0.30 |
ENSDART00000122957
|
crygm4
|
crystallin, gamma M4 |
| chr21_+_34976600 | 0.30 |
ENSDART00000191672
ENSDART00000139635 |
rbm11
|
RNA binding motif protein 11 |
| chr11_+_6159595 | 0.30 |
ENSDART00000178367
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr15_-_40065265 | 0.30 |
ENSDART00000063781
ENSDART00000142394 |
gpr55a
|
G protein-coupled receptor 55a |
Network of associatons between targets according to the STRING database.
First level regulatory network of rbpjb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 1.1 | 5.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 1.0 | 4.0 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 1.0 | 2.9 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.9 | 2.8 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.7 | 2.2 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.7 | 3.6 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.7 | 3.4 | GO:0043282 | pharyngeal muscle development(GO:0043282) |
| 0.5 | 5.4 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.4 | 3.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.4 | 3.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.3 | 1.4 | GO:0055016 | hypochord development(GO:0055016) |
| 0.3 | 1.0 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.3 | 1.7 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) positive regulation of calcium ion import(GO:0090280) |
| 0.3 | 1.9 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 0.9 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.2 | 27.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.2 | 1.6 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 4.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 1.3 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.2 | 1.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.2 | 2.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 2.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 2.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 1.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 5.8 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 1.9 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.5 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.6 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 1.0 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 3.5 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 1.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 1.9 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 1.4 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 3.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.0 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.9 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 1.9 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.2 | GO:0034138 | toll-like receptor 2 signaling pathway(GO:0034134) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.1 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.8 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 2.0 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 2.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 2.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 6.3 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.1 | 0.7 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 17.3 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.1 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.6 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 1.0 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.2 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 3.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.9 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 1.1 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 2.2 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 2.5 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 2.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 3.5 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 1.3 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.8 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 1.8 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.0 | GO:0003218 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.0 | 1.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.7 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.3 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 1.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 8.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 1.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 0.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 4.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 2.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.5 | GO:0031362 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 4.3 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 2.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 7.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 4.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.5 | 5.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.5 | 5.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 1.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 27.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 1.7 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.3 | 1.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 4.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 3.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 1.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.7 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.1 | 2.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 2.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 1.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.5 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 1.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 4.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.9 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 2.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 4.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 2.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 6.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 3.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 2.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 18.5 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 3.5 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.0 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 9.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 2.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 2.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 3.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 4.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 4.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |