Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
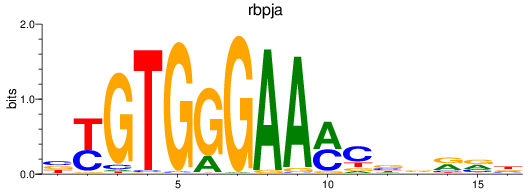
Results for rbpja
Z-value: 0.63
Transcription factors associated with rbpja
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rbpja
|
ENSDARG00000003398 | recombination signal binding protein for immunoglobulin kappa J region a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rbpja | dr11_v1_chr1_+_14454663_14454663 | 0.64 | 6.5e-02 | Click! |
Activity profile of rbpja motif
Sorted Z-values of rbpja motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_2573021 | 0.91 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr24_-_9989634 | 0.79 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr6_+_30430591 | 0.61 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr3_-_3428938 | 0.58 |
ENSDART00000179811
ENSDART00000115282 ENSDART00000192263 ENSDART00000046454 |
A2ML1 (1 of many)
|
zgc:171445 |
| chr20_+_14789148 | 0.58 |
ENSDART00000164761
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr19_+_42432625 | 0.56 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr25_+_20272145 | 0.49 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr3_-_3439150 | 0.47 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr2_-_38287987 | 0.46 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr6_-_9695294 | 0.41 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr6_-_2133737 | 0.40 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr3_+_3641429 | 0.40 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr21_-_27213166 | 0.36 |
ENSDART00000146959
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr6_+_21005725 | 0.36 |
ENSDART00000041370
|
cx44.2
|
connexin 44.2 |
| chr3_+_36646054 | 0.35 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr1_+_57348756 | 0.35 |
ENSDART00000063750
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr21_+_25221940 | 0.35 |
ENSDART00000108972
|
sycn.1
|
syncollin, tandem duplicate 1 |
| chr21_+_39673157 | 0.35 |
ENSDART00000100240
|
mrm3b
|
mitochondrial rRNA methyltransferase 3b |
| chr11_-_3535537 | 0.35 |
ENSDART00000165329
ENSDART00000009788 |
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr24_+_39027481 | 0.35 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr15_-_31147301 | 0.34 |
ENSDART00000157145
ENSDART00000155473 ENSDART00000048103 |
ksr1b
|
kinase suppressor of ras 1b |
| chr1_-_58887610 | 0.34 |
ENSDART00000180647
|
CABZ01084501.1
|
microfibril-associated glycoprotein 4-like precursor |
| chr9_+_50175366 | 0.34 |
ENSDART00000170352
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr16_-_25680666 | 0.33 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr10_+_36441124 | 0.33 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr24_+_18714212 | 0.33 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr1_-_23370395 | 0.33 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr4_-_16451375 | 0.32 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr3_+_46315016 | 0.32 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr20_+_53368611 | 0.32 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr17_+_16046132 | 0.32 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr5_-_36597612 | 0.31 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr14_+_41406321 | 0.31 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr5_-_17876709 | 0.31 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr24_-_21090447 | 0.31 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr5_-_23675222 | 0.31 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr18_-_21746421 | 0.28 |
ENSDART00000188809
|
pskh1
|
protein serine kinase H1 |
| chr19_-_18127629 | 0.28 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr23_-_45705525 | 0.28 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr19_-_18127808 | 0.28 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr8_+_15277874 | 0.28 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr2_-_32237916 | 0.28 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr18_-_18937485 | 0.27 |
ENSDART00000139015
|
si:dkey-73n10.1
|
si:dkey-73n10.1 |
| chr7_+_15329819 | 0.27 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr23_+_19790962 | 0.26 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr10_-_38468847 | 0.26 |
ENSDART00000133914
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr25_+_2263857 | 0.25 |
ENSDART00000076439
|
yars2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr20_+_14789305 | 0.25 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr1_-_31105376 | 0.25 |
ENSDART00000132466
|
ppp1r9alb
|
protein phosphatase 1 regulatory subunit 9A-like B |
| chr3_-_41995321 | 0.24 |
ENSDART00000192277
|
ttyh3a
|
tweety family member 3a |
| chr7_+_29890292 | 0.24 |
ENSDART00000170403
ENSDART00000168600 |
tln2a
|
talin 2a |
| chr16_+_2905150 | 0.24 |
ENSDART00000109980
|
lars2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr6_-_53144336 | 0.23 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr6_+_3716666 | 0.23 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr24_+_24170914 | 0.23 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr7_+_39054478 | 0.23 |
ENSDART00000173825
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr17_+_5061135 | 0.22 |
ENSDART00000064313
|
cdc5l
|
CDC5 cell division cycle 5-like (S. pombe) |
| chr14_+_32926385 | 0.22 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr5_+_61361815 | 0.21 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr12_+_1286642 | 0.21 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr6_-_55423220 | 0.21 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr11_-_16394971 | 0.21 |
ENSDART00000180981
ENSDART00000179925 |
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr18_-_17075098 | 0.21 |
ENSDART00000042496
ENSDART00000192284 ENSDART00000180307 |
tango6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr17_-_5610514 | 0.20 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr21_+_43172506 | 0.20 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr19_+_43341424 | 0.19 |
ENSDART00000134815
|
sesn2
|
sestrin 2 |
| chr25_+_16646113 | 0.19 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr8_-_36554675 | 0.19 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr24_+_20658760 | 0.19 |
ENSDART00000188362
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr16_+_8139515 | 0.18 |
ENSDART00000193303
ENSDART00000139432 |
pomgnt2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr6_+_58522557 | 0.18 |
ENSDART00000128062
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr7_+_15324830 | 0.18 |
ENSDART00000189088
|
mespaa
|
mesoderm posterior aa |
| chr6_+_58522738 | 0.18 |
ENSDART00000157327
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr25_+_34915762 | 0.18 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr23_-_478201 | 0.18 |
ENSDART00000140749
|
si:ch73-181d5.4
|
si:ch73-181d5.4 |
| chr13_-_24874950 | 0.17 |
ENSDART00000077775
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr3_+_35608385 | 0.17 |
ENSDART00000193219
ENSDART00000132703 |
traf7
|
TNF receptor-associated factor 7 |
| chr19_+_3842891 | 0.17 |
ENSDART00000159043
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr18_-_5209258 | 0.17 |
ENSDART00000183109
|
CABZ01080601.1
|
|
| chr21_+_25226558 | 0.17 |
ENSDART00000168480
|
sycn.2
|
syncollin, tandem duplicate 2 |
| chr1_-_54100988 | 0.17 |
ENSDART00000192662
|
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr3_-_53092509 | 0.17 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr16_-_44945224 | 0.17 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr4_+_55778679 | 0.17 |
ENSDART00000183009
|
CT583728.19
|
|
| chr4_+_55794876 | 0.17 |
ENSDART00000189043
|
CT583728.17
|
|
| chr4_+_55810436 | 0.17 |
ENSDART00000182875
|
CT583728.16
|
|
| chr4_-_68568233 | 0.17 |
ENSDART00000184284
|
BX548011.1
|
|
| chr20_-_54075136 | 0.17 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr20_+_23501535 | 0.16 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr21_-_32082130 | 0.16 |
ENSDART00000003978
ENSDART00000182050 |
mat2b
|
methionine adenosyltransferase II, beta |
| chr20_-_9199721 | 0.16 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr8_+_36554816 | 0.16 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr5_+_20319519 | 0.16 |
ENSDART00000004217
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr19_+_13994563 | 0.16 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr11_-_26701611 | 0.16 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr12_+_32368574 | 0.16 |
ENSDART00000086389
|
ANKFN1
|
si:ch211-277e21.2 |
| chr5_-_37997774 | 0.16 |
ENSDART00000139616
ENSDART00000167694 |
si:dkey-111e8.1
|
si:dkey-111e8.1 |
| chr19_-_44801918 | 0.15 |
ENSDART00000108507
|
CSMD3 (1 of many)
|
si:ch211-233f16.1 |
| chr18_-_46369516 | 0.15 |
ENSDART00000018163
|
irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr2_-_38282079 | 0.15 |
ENSDART00000145808
|
rnf212b
|
si:ch211-10e2.1 |
| chr18_+_18612388 | 0.15 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr8_+_12925385 | 0.15 |
ENSDART00000085377
|
KIF2A
|
zgc:103670 |
| chr22_-_26274177 | 0.15 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr7_-_41726657 | 0.15 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr18_-_18587745 | 0.14 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr18_-_6824518 | 0.14 |
ENSDART00000141132
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr17_+_45472608 | 0.14 |
ENSDART00000103493
ENSDART00000183474 |
zgc:110783
|
zgc:110783 |
| chr11_-_23322182 | 0.14 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr1_+_45839927 | 0.13 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr21_+_45268112 | 0.13 |
ENSDART00000157136
|
tcf7
|
transcription factor 7 |
| chr2_-_898899 | 0.13 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr7_+_24496894 | 0.13 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr19_+_7001170 | 0.13 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr25_+_245018 | 0.13 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr7_-_26462831 | 0.13 |
ENSDART00000113543
|
mblac1
|
metallo-beta-lactamase domain containing 1 |
| chr13_+_21919786 | 0.13 |
ENSDART00000182440
|
ndst2a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2a |
| chr5_-_32338866 | 0.13 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr2_+_8112449 | 0.13 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr5_+_65040228 | 0.12 |
ENSDART00000164278
|
pmpca
|
peptidase (mitochondrial processing) alpha |
| chr15_-_28908027 | 0.12 |
ENSDART00000182790
ENSDART00000192461 |
eml2
|
echinoderm microtubule associated protein like 2 |
| chr3_+_2669813 | 0.12 |
ENSDART00000014205
|
CR388047.1
|
|
| chr17_+_11372531 | 0.12 |
ENSDART00000130975
ENSDART00000149366 |
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr24_+_20658942 | 0.12 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr3_+_31058464 | 0.12 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr17_+_44756247 | 0.12 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr12_+_25223843 | 0.11 |
ENSDART00000077180
ENSDART00000127454 ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr8_+_23147609 | 0.11 |
ENSDART00000180284
|
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr19_+_40248697 | 0.11 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr17_+_22381215 | 0.11 |
ENSDART00000162670
ENSDART00000128875 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr3_+_10637330 | 0.11 |
ENSDART00000129257
|
tmem220
|
transmembrane protein 220 |
| chr20_-_39596338 | 0.11 |
ENSDART00000023531
|
hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr19_+_41520892 | 0.11 |
ENSDART00000182218
ENSDART00000115123 |
crtc2
|
CREB regulated transcription coactivator 2 |
| chr2_+_44781185 | 0.11 |
ENSDART00000187103
ENSDART00000147292 ENSDART00000098123 |
ece2a
|
endothelin converting enzyme 2a |
| chr10_+_573667 | 0.11 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr2_-_43850897 | 0.11 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr25_+_34915576 | 0.11 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr23_+_17839187 | 0.10 |
ENSDART00000104647
|
prim1
|
DNA primase subunit 1 |
| chr9_+_17984358 | 0.10 |
ENSDART00000192399
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr21_-_2209012 | 0.10 |
ENSDART00000158345
|
zgc:162971
|
zgc:162971 |
| chr16_-_30885838 | 0.09 |
ENSDART00000131356
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr22_-_506522 | 0.09 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr4_-_61406364 | 0.09 |
ENSDART00000142592
|
znf1021
|
zinc finger protein 1021 |
| chr2_+_57504073 | 0.09 |
ENSDART00000129811
|
onecut3b
|
one cut homeobox 3b |
| chr15_-_28907709 | 0.09 |
ENSDART00000017268
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr6_-_52235118 | 0.09 |
ENSDART00000191243
|
tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr11_-_3613558 | 0.09 |
ENSDART00000163578
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr17_+_43659940 | 0.09 |
ENSDART00000145738
ENSDART00000075619 |
adob
|
2-aminoethanethiol (cysteamine) dioxygenase b |
| chr3_-_7436932 | 0.09 |
ENSDART00000163501
|
CR847850.1
|
|
| chr17_-_45254585 | 0.08 |
ENSDART00000185507
ENSDART00000172080 |
ttbk2a
|
tau tubulin kinase 2a |
| chr1_-_47071979 | 0.08 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr5_+_27137473 | 0.08 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr15_-_25435085 | 0.08 |
ENSDART00000112079
|
tlcd2
|
TLC domain containing 2 |
| chr3_-_27784457 | 0.08 |
ENSDART00000019098
|
dnase1
|
deoxyribonuclease I |
| chr3_+_32862730 | 0.08 |
ENSDART00000144939
ENSDART00000125126 ENSDART00000144531 ENSDART00000141717 ENSDART00000137599 |
zgc:162613
|
zgc:162613 |
| chr25_+_36045072 | 0.08 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr4_-_74367912 | 0.08 |
ENSDART00000174199
ENSDART00000165257 |
ptprb
|
protein tyrosine phosphatase, receptor type, b |
| chr12_-_19862912 | 0.08 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr6_-_30683637 | 0.08 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr8_-_46386024 | 0.08 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr22_+_3238474 | 0.07 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr14_-_4321874 | 0.07 |
ENSDART00000042672
|
guf1
|
GUF1 homolog, GTPase |
| chr14_+_29775602 | 0.07 |
ENSDART00000125589
|
zgc:153146
|
zgc:153146 |
| chr2_-_1622641 | 0.07 |
ENSDART00000082143
|
prkacbb
|
protein kinase, cAMP-dependent, catalytic, beta b |
| chr23_-_36003282 | 0.07 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr7_-_17712665 | 0.07 |
ENSDART00000149047
|
men1
|
multiple endocrine neoplasia I |
| chr14_+_743346 | 0.07 |
ENSDART00000110511
|
klb
|
klotho beta |
| chr18_-_50676567 | 0.07 |
ENSDART00000172264
|
ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr23_-_14216506 | 0.07 |
ENSDART00000019620
|
ddx23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr5_-_6567464 | 0.07 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr21_-_22720381 | 0.07 |
ENSDART00000130789
|
c1qc
|
complement component 1, q subcomponent, C chain |
| chr11_-_27537593 | 0.07 |
ENSDART00000173444
ENSDART00000172895 ENSDART00000088177 |
ptpdc1a
|
protein tyrosine phosphatase domain containing 1a |
| chr5_+_41143563 | 0.07 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr23_-_9855627 | 0.07 |
ENSDART00000180159
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr17_-_31719071 | 0.07 |
ENSDART00000136199
|
dtd2
|
D-tyrosyl-tRNA deacylase 2 |
| chr16_-_30901159 | 0.07 |
ENSDART00000180313
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr3_-_36272670 | 0.07 |
ENSDART00000141638
|
prkar1aa
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) a |
| chr4_+_40329524 | 0.07 |
ENSDART00000165722
|
znf993
|
zinc finger protein 993 |
| chr13_-_49819027 | 0.06 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr8_+_23147218 | 0.06 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr3_-_36750068 | 0.06 |
ENSDART00000173388
|
abcc6b.1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 1 |
| chr14_-_7207961 | 0.06 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr11_-_8269227 | 0.06 |
ENSDART00000127202
ENSDART00000158079 ENSDART00000158748 |
si:cabz01021067.1
pimr202
|
si:cabz01021067.1 Pim proto-oncogene, serine/threonine kinase, related 202 |
| chr3_+_17616201 | 0.06 |
ENSDART00000156775
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr3_-_31158382 | 0.06 |
ENSDART00000076764
ENSDART00000076796 |
smg1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr1_-_45889820 | 0.06 |
ENSDART00000144735
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr23_-_5783421 | 0.06 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr15_+_31481939 | 0.06 |
ENSDART00000134306
|
or102-5
|
odorant receptor, family C, subfamily 102, member 5 |
| chr19_-_1023051 | 0.06 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr17_-_6508406 | 0.06 |
ENSDART00000002778
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr20_-_8110672 | 0.06 |
ENSDART00000113993
|
si:ch211-232i5.1
|
si:ch211-232i5.1 |
| chr4_-_41269844 | 0.05 |
ENSDART00000186177
|
CR388165.2
|
|
| chr7_+_61184104 | 0.05 |
ENSDART00000110671
|
zgc:194930
|
zgc:194930 |
| chr21_+_26536950 | 0.05 |
ENSDART00000146315
|
stx5al
|
syntaxin 5A, like |
| chr21_+_37436907 | 0.05 |
ENSDART00000182611
ENSDART00000076328 |
pgrmc1
|
progesterone receptor membrane component 1 |
| chr23_-_32092443 | 0.05 |
ENSDART00000133688
|
letmd1
|
LETM1 domain containing 1 |
| chr11_-_20956309 | 0.04 |
ENSDART00000188659
|
CABZ01008739.1
|
|
| chr12_+_532019 | 0.04 |
ENSDART00000152272
|
si:dkey-94l16.4
|
si:dkey-94l16.4 |
| chr4_+_45441669 | 0.04 |
ENSDART00000150745
|
si:ch211-162i8.7
|
si:ch211-162i8.7 |
| chr7_-_50410524 | 0.04 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr6_+_29860776 | 0.04 |
ENSDART00000028406
|
dlg1
|
discs, large homolog 1 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of rbpja
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 1.4 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.3 | GO:1904867 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.3 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.6 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.1 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.2 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.3 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.6 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.2 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.1 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.2 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.1 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.1 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:1990077 | primosome complex(GO:1990077) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |