Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
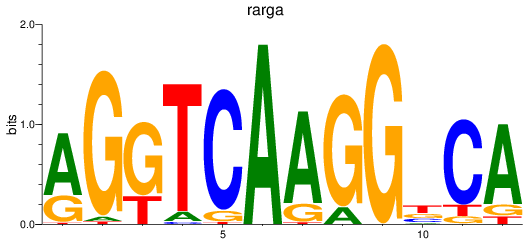
Results for rarga
Z-value: 0.90
Transcription factors associated with rarga
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rarga
|
ENSDARG00000034117 | retinoic acid receptor gamma a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rarga | dr11_v1_chr23_+_35918530_35918530 | 0.86 | 2.8e-03 | Click! |
Activity profile of rarga motif
Sorted Z-values of rarga motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_42234484 | 1.83 |
ENSDART00000132617
ENSDART00000136690 ENSDART00000141358 |
apom
|
apolipoprotein M |
| chr19_+_40856534 | 1.75 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr4_-_17409533 | 1.52 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr7_-_35708450 | 1.31 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr4_+_12031958 | 1.25 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr5_-_41531629 | 1.25 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr19_+_40856807 | 1.24 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr13_-_22862133 | 1.19 |
ENSDART00000138563
|
pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr6_+_52804267 | 1.15 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr21_-_551014 | 1.13 |
ENSDART00000099252
|
vimr1
|
vimentin-related 1 |
| chr2_+_27010439 | 1.12 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr24_-_17047918 | 1.00 |
ENSDART00000020204
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr6_-_20952187 | 0.95 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr19_+_5072918 | 0.94 |
ENSDART00000037126
|
eno2
|
enolase 2 |
| chr14_+_6159162 | 0.94 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr20_+_16750177 | 0.92 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr6_-_15653494 | 0.91 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr9_-_1970071 | 0.89 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr20_+_31269778 | 0.88 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr10_+_37927100 | 0.87 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr11_-_24681292 | 0.86 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr19_-_30404096 | 0.85 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr17_+_7595356 | 0.85 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr14_-_33454595 | 0.81 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr3_-_30685401 | 0.80 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr12_-_30583668 | 0.80 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr14_-_5678457 | 0.78 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr7_-_27033080 | 0.78 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr9_-_22834860 | 0.78 |
ENSDART00000146486
|
neb
|
nebulin |
| chr16_+_48753664 | 0.72 |
ENSDART00000155148
|
si:ch73-31d8.2
|
si:ch73-31d8.2 |
| chr10_-_27046639 | 0.71 |
ENSDART00000041841
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr13_+_22264914 | 0.69 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr19_-_30403922 | 0.68 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr6_+_52235441 | 0.67 |
ENSDART00000056319
|
cox6c
|
cytochrome c oxidase subunit VIc |
| chr24_-_17029374 | 0.67 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr5_-_64168415 | 0.67 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr18_-_22753637 | 0.64 |
ENSDART00000181589
ENSDART00000009912 |
hsf4
|
heat shock transcription factor 4 |
| chr7_-_56793739 | 0.63 |
ENSDART00000082842
|
si:ch211-146m13.3
|
si:ch211-146m13.3 |
| chr22_-_17595310 | 0.61 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr20_+_572037 | 0.58 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr9_+_31795343 | 0.57 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr4_-_17629444 | 0.57 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr6_-_49898881 | 0.57 |
ENSDART00000150204
|
atp5f1e
|
ATP synthase F1 subunit epsilon |
| chr2_+_24536762 | 0.57 |
ENSDART00000144149
|
angptl4
|
angiopoietin-like 4 |
| chr11_+_37201483 | 0.56 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr6_+_41099787 | 0.56 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr7_+_20512419 | 0.55 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr8_+_19514294 | 0.53 |
ENSDART00000170622
|
si:ch73-281k2.5
|
si:ch73-281k2.5 |
| chr8_+_1769475 | 0.53 |
ENSDART00000079073
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr21_-_5879897 | 0.53 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr7_-_58729894 | 0.53 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr13_+_31108334 | 0.53 |
ENSDART00000142245
|
arhgap22
|
Rho GTPase activating protein 22 |
| chr10_-_35220285 | 0.53 |
ENSDART00000180439
|
ypel2a
|
yippee-like 2a |
| chr19_+_30387999 | 0.52 |
ENSDART00000145396
|
tspan13b
|
tetraspanin 13b |
| chr13_-_22843562 | 0.51 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr8_-_25329967 | 0.50 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr9_-_1986014 | 0.49 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr5_-_35301800 | 0.48 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr19_-_103289 | 0.47 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr3_+_29458517 | 0.47 |
ENSDART00000134258
|
grap2a
|
GRB2-related adaptor protein 2a |
| chr13_+_22480496 | 0.46 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr23_+_36087219 | 0.46 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr22_+_27284462 | 0.46 |
ENSDART00000164660
|
si:ch73-103b2.1
|
si:ch73-103b2.1 |
| chr13_+_15182149 | 0.46 |
ENSDART00000193644
ENSDART00000134421 ENSDART00000086281 |
mavs
|
mitochondrial antiviral signaling protein |
| chr5_-_55395964 | 0.45 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr23_+_24789205 | 0.44 |
ENSDART00000088697
|
olfml3a
|
olfactomedin-like 3a |
| chr21_+_19635486 | 0.44 |
ENSDART00000185736
|
fgf10a
|
fibroblast growth factor 10a |
| chr20_+_26095530 | 0.44 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr18_-_1185772 | 0.44 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr3_-_18792492 | 0.41 |
ENSDART00000134208
ENSDART00000034373 |
hagh
|
hydroxyacylglutathione hydrolase |
| chr9_-_34871900 | 0.41 |
ENSDART00000026378
|
slc25a6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr5_+_30179010 | 0.40 |
ENSDART00000134624
|
adamts15a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15a |
| chr6_+_48618512 | 0.40 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr1_+_54043563 | 0.39 |
ENSDART00000149760
|
triobpa
|
TRIO and F-actin binding protein a |
| chr5_+_20693724 | 0.39 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr2_-_24289641 | 0.38 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr13_+_22480857 | 0.38 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr22_-_18779232 | 0.37 |
ENSDART00000186726
|
atp5f1d
|
ATP synthase F1 subunit delta |
| chr16_-_17197546 | 0.37 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr22_-_18778988 | 0.37 |
ENSDART00000019235
|
atp5f1d
|
ATP synthase F1 subunit delta |
| chr18_-_14937211 | 0.36 |
ENSDART00000141893
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr15_+_19682013 | 0.35 |
ENSDART00000127368
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr10_+_29260096 | 0.35 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr17_+_23770848 | 0.35 |
ENSDART00000079646
|
kcnk18
|
potassium channel, subfamily K, member 18 |
| chr16_+_27345383 | 0.34 |
ENSDART00000078250
ENSDART00000162857 |
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr2_+_38264964 | 0.33 |
ENSDART00000182068
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr1_-_22726233 | 0.33 |
ENSDART00000140920
|
prom1b
|
prominin 1 b |
| chr21_-_43117327 | 0.32 |
ENSDART00000122352
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr24_-_14711597 | 0.32 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr24_-_25363815 | 0.31 |
ENSDART00000186814
|
ptchd1
|
patched domain containing 1 |
| chr21_+_44112914 | 0.30 |
ENSDART00000062836
|
fgf1b
|
fibroblast growth factor 1b |
| chr14_-_42997145 | 0.30 |
ENSDART00000172801
|
pcdh10b
|
protocadherin 10b |
| chr19_-_703898 | 0.30 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr16_-_7228276 | 0.30 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr6_+_18569453 | 0.30 |
ENSDART00000171338
|
rhot1b
|
ras homolog family member T1 |
| chr14_-_36412473 | 0.29 |
ENSDART00000128244
ENSDART00000138376 |
asb5a
|
ankyrin repeat and SOCS box containing 5a |
| chr3_-_62393449 | 0.29 |
ENSDART00000101870
ENSDART00000140782 ENSDART00000181704 |
proza
|
protein Z, vitamin K-dependent plasma glycoprotein a |
| chr7_-_31618166 | 0.29 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr15_+_3284416 | 0.28 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr23_-_19953089 | 0.28 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr10_-_24648228 | 0.28 |
ENSDART00000081834
ENSDART00000132830 |
stoml3b
|
stomatin (EPB72)-like 3b |
| chr15_-_2632891 | 0.26 |
ENSDART00000081840
|
cldnj
|
claudin j |
| chr6_-_10835849 | 0.26 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr15_-_20933574 | 0.26 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr16_+_16978424 | 0.25 |
ENSDART00000143128
|
rpl18
|
ribosomal protein L18 |
| chr7_+_15266093 | 0.25 |
ENSDART00000124676
|
sv2ba
|
synaptic vesicle glycoprotein 2Ba |
| chr15_+_1004680 | 0.25 |
ENSDART00000157310
|
si:dkey-77f5.8
|
si:dkey-77f5.8 |
| chr20_-_25709247 | 0.25 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr2_-_31634978 | 0.24 |
ENSDART00000135668
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr8_+_668184 | 0.24 |
ENSDART00000183788
|
rnf165b
|
ring finger protein 165b |
| chr7_-_50883433 | 0.24 |
ENSDART00000174314
|
PAQR9
|
progestin and adipoQ receptor family member 9 |
| chr10_-_15963903 | 0.24 |
ENSDART00000142357
|
si:dkey-3h23.3
|
si:dkey-3h23.3 |
| chr15_-_31514818 | 0.24 |
ENSDART00000153978
|
hmgb1b
|
high mobility group box 1b |
| chr21_-_20939488 | 0.23 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr10_-_43294933 | 0.23 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr17_-_15498275 | 0.23 |
ENSDART00000156905
ENSDART00000080661 |
si:ch211-266g18.10
|
si:ch211-266g18.10 |
| chr2_+_7818368 | 0.23 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr24_+_3328354 | 0.23 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr1_-_46632948 | 0.22 |
ENSDART00000148893
ENSDART00000053232 |
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr20_+_192170 | 0.22 |
ENSDART00000189675
|
cx28.8
|
connexin 28.8 |
| chr10_-_16028082 | 0.22 |
ENSDART00000122540
|
aldh7a1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr21_+_5801105 | 0.22 |
ENSDART00000151225
ENSDART00000184487 |
ccng2
|
cyclin G2 |
| chr18_-_34143189 | 0.22 |
ENSDART00000079341
|
plch1
|
phospholipase C, eta 1 |
| chr1_-_9109699 | 0.21 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr20_+_11800063 | 0.21 |
ENSDART00000152230
|
si:ch211-155o21.4
|
si:ch211-155o21.4 |
| chr10_-_4980150 | 0.21 |
ENSDART00000093228
|
mat2al
|
methionine adenosyltransferase II, alpha-like |
| chr2_+_33382648 | 0.21 |
ENSDART00000137207
ENSDART00000098831 |
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr5_+_42467867 | 0.20 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr2_+_51028269 | 0.20 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr23_+_42336084 | 0.20 |
ENSDART00000158959
ENSDART00000161812 |
cyp2aa7
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr19_-_42424599 | 0.20 |
ENSDART00000077042
|
zgc:153441
|
zgc:153441 |
| chr17_-_2945592 | 0.19 |
ENSDART00000174899
|
C14orf132
|
chromosome 14 open reading frame 132 |
| chr5_-_71838520 | 0.19 |
ENSDART00000174396
|
CU927890.1
|
|
| chr14_-_44841503 | 0.19 |
ENSDART00000179114
|
GRXCR1
|
si:dkey-109l4.6 |
| chr21_-_29100110 | 0.19 |
ENSDART00000142598
|
timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr10_-_39321367 | 0.19 |
ENSDART00000129647
|
smtlb
|
somatolactin beta |
| chr9_+_24095677 | 0.19 |
ENSDART00000150443
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr16_-_17116499 | 0.19 |
ENSDART00000138983
|
si:dkey-260g12.1
|
si:dkey-260g12.1 |
| chr10_+_34623183 | 0.18 |
ENSDART00000114630
|
nbeaa
|
neurobeachin a |
| chr9_-_48281941 | 0.18 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr15_+_24644251 | 0.17 |
ENSDART00000181660
|
smtnl
|
smoothelin, like |
| chr17_-_43287290 | 0.17 |
ENSDART00000156885
|
EML5
|
si:dkey-1f12.3 |
| chr7_+_42206543 | 0.17 |
ENSDART00000112543
|
phkb
|
phosphorylase kinase, beta |
| chr25_+_3326885 | 0.17 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr4_-_23839789 | 0.16 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr11_+_3578543 | 0.16 |
ENSDART00000191015
|
si:dkey-33m11.8
|
si:dkey-33m11.8 |
| chr15_+_19681718 | 0.16 |
ENSDART00000164803
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr16_-_40727455 | 0.16 |
ENSDART00000162331
|
si:dkey-22o22.2
|
si:dkey-22o22.2 |
| chr18_-_226800 | 0.16 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr2_-_27774783 | 0.16 |
ENSDART00000161864
|
XKR4
|
zgc:123035 |
| chr21_+_40225915 | 0.16 |
ENSDART00000048475
ENSDART00000174122 |
or115-12
|
odorant receptor, family F, subfamily 115, member 12 |
| chr6_+_11990733 | 0.16 |
ENSDART00000151075
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr5_+_1079423 | 0.16 |
ENSDART00000172231
|
si:zfos-128g4.2
|
si:zfos-128g4.2 |
| chr25_+_3327071 | 0.16 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr5_+_69868911 | 0.15 |
ENSDART00000014649
ENSDART00000188215 ENSDART00000167385 |
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr4_-_13567387 | 0.15 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr24_+_28528000 | 0.15 |
ENSDART00000155924
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr12_-_7806007 | 0.15 |
ENSDART00000190359
|
ank3b
|
ankyrin 3b |
| chr8_+_20679759 | 0.15 |
ENSDART00000088668
|
nfic
|
nuclear factor I/C |
| chr8_-_50287949 | 0.15 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr14_+_30795559 | 0.15 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr2_-_53592532 | 0.14 |
ENSDART00000184066
|
ccl25a
|
chemokine (C-C motif) ligand 25a |
| chr3_+_41922114 | 0.14 |
ENSDART00000138280
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr9_-_303658 | 0.14 |
ENSDART00000160338
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr14_-_44841335 | 0.14 |
ENSDART00000173011
|
GRXCR1
|
si:dkey-109l4.6 |
| chr21_+_30355767 | 0.14 |
ENSDART00000189948
|
CR749164.1
|
|
| chr13_+_24579108 | 0.13 |
ENSDART00000001830
|
echs1
|
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr8_+_22851472 | 0.13 |
ENSDART00000041564
|
cacna1fb
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr2_+_24868010 | 0.13 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr14_+_51015205 | 0.13 |
ENSDART00000074216
|
higd2a
|
HIG1 hypoxia inducible domain family, member 2A |
| chr21_+_28724099 | 0.13 |
ENSDART00000138017
|
puraa
|
purine-rich element binding protein Aa |
| chr7_+_26100024 | 0.13 |
ENSDART00000173726
|
si:ch211-196f2.3
|
si:ch211-196f2.3 |
| chr22_-_4780010 | 0.13 |
ENSDART00000192844
|
si:ch73-256j6.5
|
si:ch73-256j6.5 |
| chr4_-_73825089 | 0.12 |
ENSDART00000174207
|
si:dkey-262g12.12
|
si:dkey-262g12.12 |
| chr20_+_6142433 | 0.12 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr6_+_28054639 | 0.12 |
ENSDART00000187478
ENSDART00000189194 |
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr7_+_52135791 | 0.12 |
ENSDART00000098705
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr9_-_44939104 | 0.12 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr8_+_49778486 | 0.11 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr21_-_22831388 | 0.11 |
ENSDART00000151040
|
angptl5
|
angiopoietin-like 5 |
| chr17_-_26610814 | 0.11 |
ENSDART00000133402
ENSDART00000016608 |
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr14_+_16034447 | 0.11 |
ENSDART00000161348
|
prelid1a
|
PRELI domain containing 1a |
| chr2_-_34483597 | 0.11 |
ENSDART00000133224
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr21_-_30658509 | 0.11 |
ENSDART00000139764
|
si:dkey-22f5.9
|
si:dkey-22f5.9 |
| chr13_-_15994419 | 0.11 |
ENSDART00000079724
ENSDART00000042377 ENSDART00000046079 ENSDART00000050481 ENSDART00000016430 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr2_-_47431205 | 0.11 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr8_+_22472584 | 0.10 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr20_-_14665002 | 0.10 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr6_-_10912424 | 0.10 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr1_+_58282449 | 0.10 |
ENSDART00000131475
|
si:dkey-222h21.7
|
si:dkey-222h21.7 |
| chr12_-_13318944 | 0.10 |
ENSDART00000152201
ENSDART00000041394 |
emc9
|
ER membrane protein complex subunit 9 |
| chr2_-_20120904 | 0.10 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr4_+_2092683 | 0.10 |
ENSDART00000067423
|
frs2a
|
fibroblast growth factor receptor substrate 2a |
| chr11_-_36263886 | 0.10 |
ENSDART00000140397
|
nfya
|
nuclear transcription factor Y, alpha |
| chr7_-_41338923 | 0.10 |
ENSDART00000099138
|
ncf2
|
neutrophil cytosolic factor 2 |
| chr1_+_57331813 | 0.10 |
ENSDART00000152440
ENSDART00000062841 |
epn3b
|
epsin 3b |
| chr18_-_40508528 | 0.09 |
ENSDART00000185249
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr3_+_21189766 | 0.09 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr20_+_33534038 | 0.09 |
ENSDART00000029206
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr25_+_25766033 | 0.09 |
ENSDART00000103638
ENSDART00000039952 |
idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of rarga
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.6 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.2 | 0.9 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 1.3 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 1.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 1.5 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.9 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.5 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.1 | 0.7 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.4 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.4 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 1.0 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.1 | 0.5 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.1 | 0.5 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.8 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.6 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 1.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.3 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 0.2 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.0 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 1.1 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.5 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.3 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.4 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.1 | GO:0002456 | T cell cytokine production(GO:0002369) T cell mediated immunity(GO:0002456) |
| 0.0 | 0.3 | GO:0030324 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 1.2 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.6 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.3 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.4 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 1.0 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.3 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.8 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.2 | 3.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 0.9 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.1 | 0.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 1.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 1.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.2 | 1.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 1.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 3.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.6 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.9 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 0.4 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 1.0 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.7 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.2 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.1 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.5 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.1 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |