Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
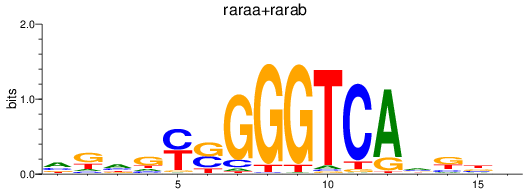
Results for raraa+rarab
Z-value: 0.78
Transcription factors associated with raraa+rarab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rarab
|
ENSDARG00000034893 | retinoic acid receptor, alpha b |
|
raraa
|
ENSDARG00000056783 | retinoic acid receptor, alpha a |
|
rarab
|
ENSDARG00000111757 | retinoic acid receptor, alpha b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rarab | dr11_v1_chr3_-_33175583_33175583 | -0.97 | 1.2e-05 | Click! |
| raraa | dr11_v1_chr12_+_11080776_11080776 | 0.79 | 1.1e-02 | Click! |
Activity profile of raraa+rarab motif
Sorted Z-values of raraa+rarab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_28445052 | 2.79 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr19_+_19762183 | 1.26 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr13_+_24834199 | 1.14 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr1_+_59538755 | 1.11 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr14_-_51855047 | 1.01 |
ENSDART00000088912
|
cplx1
|
complexin 1 |
| chr6_-_8277221 | 1.00 |
ENSDART00000053869
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr17_+_53311618 | 0.95 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr5_-_64823750 | 0.94 |
ENSDART00000140305
|
lix1
|
limb and CNS expressed 1 |
| chr2_+_52847049 | 0.94 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr22_+_38173960 | 0.92 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr8_+_37168570 | 0.86 |
ENSDART00000098636
|
csf1b
|
colony stimulating factor 1b (macrophage) |
| chr4_-_191736 | 0.76 |
ENSDART00000169187
ENSDART00000192054 |
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr4_+_1530287 | 0.75 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr14_-_2001386 | 0.74 |
ENSDART00000170346
ENSDART00000163975 |
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr14_-_36862745 | 0.73 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr22_+_30009926 | 0.73 |
ENSDART00000142529
|
si:dkey-286j15.1
|
si:dkey-286j15.1 |
| chr14_-_52552875 | 0.66 |
ENSDART00000170107
|
ppp2r2ba
|
protein phosphatase 2, regulatory subunit B, beta a |
| chr17_+_14886828 | 0.66 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr5_+_64856666 | 0.57 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr16_+_7242610 | 0.55 |
ENSDART00000081477
|
sri
|
sorcin |
| chr2_+_3696038 | 0.51 |
ENSDART00000150499
ENSDART00000136906 ENSDART00000164152 ENSDART00000128514 ENSDART00000108964 |
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr4_+_3980247 | 0.49 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr22_+_20208185 | 0.46 |
ENSDART00000142748
|
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr16_-_22194400 | 0.46 |
ENSDART00000186042
|
il6r
|
interleukin 6 receptor |
| chr23_+_25305431 | 0.43 |
ENSDART00000143291
|
RPL41
|
si:dkey-151g10.6 |
| chr18_+_8340886 | 0.41 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr24_+_21973929 | 0.40 |
ENSDART00000042495
|
sat1b
|
spermidine/spermine N1-acetyltransferase 1b |
| chr12_+_2677303 | 0.38 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr17_+_14711765 | 0.37 |
ENSDART00000012889
|
cx28.6
|
connexin 28.6 |
| chr21_-_32284532 | 0.35 |
ENSDART00000190676
|
clk4b
|
CDC-like kinase 4b |
| chr21_-_43482426 | 0.34 |
ENSDART00000192901
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr17_+_33340675 | 0.32 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr23_+_25822742 | 0.32 |
ENSDART00000184436
|
r3hdml
|
R3H domain containing-like |
| chr24_-_36727922 | 0.29 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr23_-_3703569 | 0.28 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr25_+_1549838 | 0.26 |
ENSDART00000115001
|
avpr1aa
|
arginine vasopressin receptor 1Aa |
| chr14_+_94946 | 0.24 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr3_+_57268363 | 0.23 |
ENSDART00000180725
|
TMEM235 (1 of many)
|
transmembrane protein 235 |
| chr11_+_25111846 | 0.22 |
ENSDART00000128705
ENSDART00000190058 |
ndrg3a
|
ndrg family member 3a |
| chr2_+_58841181 | 0.22 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr17_+_53311243 | 0.17 |
ENSDART00000160241
ENSDART00000160009 ENSDART00000162239 |
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr11_-_34147205 | 0.17 |
ENSDART00000173216
|
atp13a3
|
ATPase 13A3 |
| chr22_+_21324398 | 0.17 |
ENSDART00000168509
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr18_+_3338228 | 0.13 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr1_+_59154521 | 0.12 |
ENSDART00000130089
ENSDART00000152456 |
soul5l
|
heme-binding protein soul5, like |
| chr18_-_36066087 | 0.12 |
ENSDART00000059352
ENSDART00000145177 |
exosc5
|
exosome component 5 |
| chr18_+_7073130 | 0.11 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr18_+_19842569 | 0.10 |
ENSDART00000147470
|
iqch
|
IQ motif containing H |
| chr17_+_46739693 | 0.09 |
ENSDART00000097810
|
pimr22
|
Pim proto-oncogene, serine/threonine kinase, related 22 |
| chr4_+_73215536 | 0.08 |
ENSDART00000174290
|
LO018260.2
|
Danio rerio protein NLRC3-like (LOC101883187), mRNA. |
| chr22_-_31788170 | 0.06 |
ENSDART00000170925
|
pimr207
|
Pim proto-oncogene, serine/threonine kinase, related 207 |
| chr15_+_1199407 | 0.05 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr8_-_28342313 | 0.03 |
ENSDART00000005583
|
kdm5bb
|
lysine (K)-specific demethylase 5Bb |
| chr2_-_55317035 | 0.02 |
ENSDART00000169382
ENSDART00000097874 |
tpm4b
|
tropomyosin 4b |
| chr14_+_36497250 | 0.01 |
ENSDART00000184727
|
TENM3
|
si:dkey-237h12.3 |
| chr3_+_5297493 | 0.01 |
ENSDART00000138596
|
si:ch211-150d5.3
|
si:ch211-150d5.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of raraa+rarab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.2 | 1.0 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 1.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.7 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.3 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.4 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.5 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.3 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.4 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.8 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 1.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |