Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
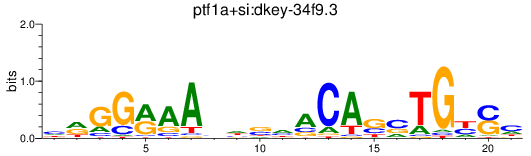
Results for ptf1a+si:dkey-34f9.3
Z-value: 1.29
Transcription factors associated with ptf1a+si:dkey-34f9.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ptf1a
|
ENSDARG00000014479 | pancreas associated transcription factor 1a |
|
si_dkey-34f9.3
|
ENSDARG00000077813 | si_dkey-34f9.3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ptf1a | dr11_v1_chr2_+_29710857_29710857 | 0.05 | 9.0e-01 | Click! |
| si:dkey-34f9.3 | dr11_v1_chr9_+_45789887_45789887 | 0.01 | 9.8e-01 | Click! |
Activity profile of ptf1a+si:dkey-34f9.3 motif
Sorted Z-values of ptf1a+si:dkey-34f9.3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_6172154 | 2.20 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr21_-_25601648 | 2.00 |
ENSDART00000042578
|
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr24_-_33756003 | 1.93 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr16_-_46572834 | 1.88 |
ENSDART00000115248
|
zgc:174938
|
zgc:174938 |
| chr15_-_46828958 | 1.87 |
ENSDART00000045110
|
chrd
|
chordin |
| chr16_-_46573211 | 1.77 |
ENSDART00000193392
|
zgc:174938
|
zgc:174938 |
| chr17_-_11015878 | 1.72 |
ENSDART00000162249
|
dact1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr1_-_59407322 | 1.57 |
ENSDART00000161661
|
si:ch211-188p14.5
|
si:ch211-188p14.5 |
| chr10_-_15048781 | 1.50 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr15_-_26636826 | 1.49 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr4_-_76488581 | 1.47 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr18_-_45736 | 1.42 |
ENSDART00000148373
ENSDART00000148950 |
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr25_-_34670413 | 1.23 |
ENSDART00000073440
|
DNAJA4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr19_-_617246 | 1.20 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr4_-_76488854 | 1.17 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr2_+_42236118 | 1.11 |
ENSDART00000114162
ENSDART00000141199 |
ftr02
|
finTRIM family, member 2 |
| chr16_+_9762261 | 1.10 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr2_-_59327299 | 1.09 |
ENSDART00000133734
|
ftr36
|
finTRIM family, member 36 |
| chr3_+_41917499 | 1.09 |
ENSDART00000028673
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr3_-_32934359 | 1.02 |
ENSDART00000124160
|
aoc2
|
amine oxidase, copper containing 2 |
| chr21_+_244503 | 0.99 |
ENSDART00000162889
|
stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr10_+_45059702 | 0.90 |
ENSDART00000166945
|
nudcd3
|
NudC domain containing 3 |
| chr10_+_506538 | 0.88 |
ENSDART00000141713
|
si:ch211-242f23.3
|
si:ch211-242f23.3 |
| chr1_+_54835131 | 0.87 |
ENSDART00000145070
|
si:ch211-196h16.4
|
si:ch211-196h16.4 |
| chr2_-_59231017 | 0.85 |
ENSDART00000143980
ENSDART00000184190 |
ftr29
|
finTRIM family, member 29 |
| chr3_+_61522759 | 0.84 |
ENSDART00000106266
|
znf1004
|
zinc finger protein 1004 |
| chr7_+_49664174 | 0.77 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr9_-_17417628 | 0.76 |
ENSDART00000060425
ENSDART00000141997 |
ftr53
|
finTRIM family, member 53 |
| chr4_-_58964138 | 0.76 |
ENSDART00000150259
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr4_-_33371634 | 0.72 |
ENSDART00000150712
|
si:dkeyp-4f2.3
|
si:dkeyp-4f2.3 |
| chr3_-_36115339 | 0.72 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr12_-_3940768 | 0.62 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr22_-_17489040 | 0.61 |
ENSDART00000141286
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr20_+_7084154 | 0.61 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr4_+_38981587 | 0.59 |
ENSDART00000142713
|
si:dkey-66k12.3
|
si:dkey-66k12.3 |
| chr4_+_37406676 | 0.58 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr12_+_7865470 | 0.56 |
ENSDART00000161683
|
BX548028.1
|
|
| chr22_-_36690742 | 0.54 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr2_-_59345920 | 0.54 |
ENSDART00000134662
|
ftr37
|
finTRIM family, member 37 |
| chr18_+_5490668 | 0.53 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr15_+_2475894 | 0.53 |
ENSDART00000035939
|
panx1a
|
pannexin 1a |
| chr22_+_1541083 | 0.52 |
ENSDART00000169338
|
si:ch211-255f4.7
|
si:ch211-255f4.7 |
| chr2_-_59178742 | 0.50 |
ENSDART00000170594
ENSDART00000109246 |
ftr95
|
finTRIM family, member 95 |
| chr18_-_1414760 | 0.50 |
ENSDART00000171881
|
PEPD
|
peptidase D |
| chr23_-_46020226 | 0.49 |
ENSDART00000160010
|
SYDE2
|
synapse defective Rho GTPase homolog 2 |
| chr4_-_41772315 | 0.48 |
ENSDART00000193109
|
znf976
|
zinc finger protein 976 |
| chr2_-_59265521 | 0.47 |
ENSDART00000146341
ENSDART00000097799 |
ftr33
|
finTRIM family, member 33 |
| chr12_+_18663154 | 0.46 |
ENSDART00000057918
|
si:ch211-147h1.4
|
si:ch211-147h1.4 |
| chr7_-_64152087 | 0.45 |
ENSDART00000165234
|
CR354398.1
|
|
| chr7_-_38644560 | 0.45 |
ENSDART00000114934
|
c6ast3
|
six-cysteine containing astacin protease 3 |
| chr9_+_28693592 | 0.44 |
ENSDART00000110198
|
zgc:162780
|
zgc:162780 |
| chr3_-_17871846 | 0.44 |
ENSDART00000074478
ENSDART00000187941 |
nkiras2
|
NFKB inhibitor interacting Ras-like 2 |
| chr3_-_58585854 | 0.43 |
ENSDART00000123546
|
CABZ01038521.1
|
|
| chr7_+_44608224 | 0.43 |
ENSDART00000005033
|
cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr15_-_2519640 | 0.42 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr7_+_44608478 | 0.42 |
ENSDART00000149981
|
cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr23_+_9353552 | 0.42 |
ENSDART00000163298
|
BX511246.1
|
|
| chr2_-_43151358 | 0.42 |
ENSDART00000061170
ENSDART00000056161 |
ftr15
|
finTRIM family, member 15 |
| chr9_+_3153717 | 0.41 |
ENSDART00000191958
|
si:zfos-979f1.2
|
si:zfos-979f1.2 |
| chr2_-_59376399 | 0.41 |
ENSDART00000137134
|
ftr38
|
finTRIM family, member 38 |
| chr2_-_42628028 | 0.40 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr25_+_36336920 | 0.40 |
ENSDART00000073402
|
hist1h2a1
|
histone cluster 1 H2A family member1 |
| chr4_-_7231480 | 0.37 |
ENSDART00000159436
|
si:ch211-170p16.1
|
si:ch211-170p16.1 |
| chr14_+_597532 | 0.37 |
ENSDART00000159805
|
LO018568.2
|
|
| chr1_-_58009216 | 0.36 |
ENSDART00000143829
|
nxnl1
|
nucleoredoxin like 1 |
| chr3_+_5504990 | 0.36 |
ENSDART00000143626
|
si:ch73-264i18.2
|
si:ch73-264i18.2 |
| chr9_+_3153466 | 0.35 |
ENSDART00000135619
|
si:zfos-979f1.2
|
si:zfos-979f1.2 |
| chr2_+_47935476 | 0.35 |
ENSDART00000140555
|
ftr26
|
finTRIM family, member 26 |
| chr2_-_47904043 | 0.35 |
ENSDART00000185328
ENSDART00000126740 |
ftr22
|
finTRIM family, member 22 |
| chr2_+_47927026 | 0.34 |
ENSDART00000143023
|
ftr25
|
finTRIM family, member 25 |
| chr24_-_6029314 | 0.33 |
ENSDART00000136155
|
ftr60
|
finTRIM family, member 60 |
| chr18_+_17537975 | 0.32 |
ENSDART00000179783
|
nup93
|
nucleoporin 93 |
| chr23_-_6865946 | 0.32 |
ENSDART00000056426
|
ftr58
|
finTRIM family, member 58 |
| chr18_-_19491162 | 0.32 |
ENSDART00000090413
|
snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr17_-_86548 | 0.31 |
ENSDART00000163657
|
grem1b
|
gremlin 1b |
| chr3_+_3545825 | 0.31 |
ENSDART00000109060
|
CR589947.1
|
|
| chr8_+_4333914 | 0.30 |
ENSDART00000137528
|
myl2b
|
myosin, light chain 2b, regulatory, cardiac, slow |
| chr8_+_16758304 | 0.30 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr3_-_34624745 | 0.29 |
ENSDART00000151091
|
tac4
|
tachykinin 4 (hemokinin) |
| chr2_-_43130812 | 0.29 |
ENSDART00000039079
ENSDART00000056162 |
ftr13
|
finTRIM family, member 13 |
| chr15_-_35134918 | 0.29 |
ENSDART00000157196
|
si:ch211-272b8.7
|
si:ch211-272b8.7 |
| chr2_-_14793343 | 0.28 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr4_-_38281909 | 0.28 |
ENSDART00000162448
|
si:ch211-229l10.9
|
si:ch211-229l10.9 |
| chr25_-_15496485 | 0.28 |
ENSDART00000140245
|
si:dkeyp-67e1.6
|
si:dkeyp-67e1.6 |
| chr3_-_61592417 | 0.27 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr20_+_38725662 | 0.26 |
ENSDART00000181873
|
uts1
|
urotensin 1 |
| chr15_-_42736433 | 0.26 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr3_-_48993580 | 0.25 |
ENSDART00000182400
|
BX769173.1
|
|
| chr3_+_18097700 | 0.25 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr11_-_36046458 | 0.25 |
ENSDART00000061520
|
brk1
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr24_+_6353394 | 0.24 |
ENSDART00000165118
|
CR352329.1
|
|
| chr23_+_4890693 | 0.24 |
ENSDART00000023537
|
tnnc1a
|
troponin C type 1a (slow) |
| chr18_+_17537344 | 0.23 |
ENSDART00000025782
|
nup93
|
nucleoporin 93 |
| chr11_-_22372072 | 0.23 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr2_+_42294944 | 0.23 |
ENSDART00000140599
ENSDART00000178811 |
ftr06
|
finTRIM family, member 6 |
| chr16_-_16152199 | 0.23 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr2_-_44368824 | 0.22 |
ENSDART00000155662
|
si:ch211-265g22.4
|
si:ch211-265g22.4 |
| chr18_-_5875433 | 0.21 |
ENSDART00000151727
|
nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr11_-_36046741 | 0.21 |
ENSDART00000184229
|
brk1
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr16_-_30932533 | 0.21 |
ENSDART00000085761
|
slc45a4
|
solute carrier family 45, member 4 |
| chr12_-_37343858 | 0.21 |
ENSDART00000153281
|
tekt3
|
tektin 3 |
| chr24_+_26088143 | 0.21 |
ENSDART00000171129
|
CR352209.1
|
|
| chr3_+_15907297 | 0.20 |
ENSDART00000139206
|
mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr18_-_18543358 | 0.20 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr1_-_59287410 | 0.20 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr3_+_62196672 | 0.20 |
ENSDART00000097312
|
sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr16_-_27676123 | 0.20 |
ENSDART00000180804
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr19_-_9662958 | 0.19 |
ENSDART00000041094
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr5_-_30704390 | 0.18 |
ENSDART00000016709
|
ift22
|
intraflagellar transport 22 homolog (Chlamydomonas) |
| chr18_-_41754020 | 0.18 |
ENSDART00000077898
|
grk7b
|
G protein-coupled receptor kinase 7b |
| chr3_-_48980319 | 0.18 |
ENSDART00000165319
|
ftr42
|
finTRIM family, member 42 |
| chr22_+_5123479 | 0.17 |
ENSDART00000111822
ENSDART00000081910 |
loxl5a
|
lysyl oxidase-like 5a |
| chr9_+_343062 | 0.17 |
ENSDART00000164012
ENSDART00000164365 |
bpifcl
|
BPI fold containing family C, like |
| chr15_-_37620167 | 0.17 |
ENSDART00000059582
|
si:ch211-137j23.8
|
si:ch211-137j23.8 |
| chr4_+_77681389 | 0.16 |
ENSDART00000099727
|
gbp4
|
guanylate binding protein 4 |
| chr7_-_38644287 | 0.16 |
ENSDART00000182307
|
c6ast3
|
six-cysteine containing astacin protease 3 |
| chr2_-_59205393 | 0.15 |
ENSDART00000056417
ENSDART00000182452 ENSDART00000141876 |
ftr30
|
finTRIM family, member 30 |
| chr17_-_30473374 | 0.14 |
ENSDART00000155021
|
si:ch211-175f11.5
|
si:ch211-175f11.5 |
| chr8_-_36554675 | 0.13 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr4_-_71983381 | 0.13 |
ENSDART00000169130
|
ftr64
|
finTRIM family, member 64 |
| chr7_+_71547747 | 0.13 |
ENSDART00000180869
|
adcyap1a
|
adenylate cyclase activating polypeptide 1a |
| chr5_+_62723233 | 0.12 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr21_+_45819662 | 0.12 |
ENSDART00000193362
ENSDART00000184255 |
pitx1
|
paired-like homeodomain 1 |
| chr20_+_53278746 | 0.11 |
ENSDART00000074249
|
gpr6
|
G protein-coupled receptor 6 |
| chr3_+_57788761 | 0.11 |
ENSDART00000149741
|
si:ch73-362i18.1
|
si:ch73-362i18.1 |
| chr8_+_54263946 | 0.10 |
ENSDART00000193119
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr23_+_5736226 | 0.09 |
ENSDART00000134527
ENSDART00000112220 |
ftr57
|
finTRIM family, member 57 |
| chr14_-_30390145 | 0.09 |
ENSDART00000045423
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr23_-_10914275 | 0.09 |
ENSDART00000112965
|
pdzrn3a
|
PDZ domain containing RING finger 3a |
| chr1_-_40123943 | 0.09 |
ENSDART00000146917
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr17_+_21477892 | 0.09 |
ENSDART00000155309
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr15_-_5212930 | 0.08 |
ENSDART00000136845
|
or128-6
|
odorant receptor, family E, subfamily 128, member 6 |
| chr3_-_4557653 | 0.08 |
ENSDART00000111305
|
ftr50
|
finTRIM family, member 50 |
| chr12_+_47122104 | 0.08 |
ENSDART00000184248
|
CABZ01088982.1
|
|
| chr13_-_638485 | 0.08 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr3_-_54524194 | 0.07 |
ENSDART00000155406
ENSDART00000111791 |
si:ch73-208g10.1
|
si:ch73-208g10.1 |
| chr13_+_9100 | 0.07 |
ENSDART00000165772
|
ppp4r3b
|
protein phosphatase 4, regulatory subunit 3B |
| chr21_-_20733615 | 0.07 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr2_+_11670270 | 0.05 |
ENSDART00000100524
|
ftr01
|
finTRIM family, member 1 |
| chr15_-_37610434 | 0.05 |
ENSDART00000156856
|
CR848763.1
|
|
| chr15_-_41523225 | 0.05 |
ENSDART00000141202
ENSDART00000099290 ENSDART00000170395 ENSDART00000099437 |
si:dkey-230i18.2
si:dkey-121n8.7
nlrc9
|
si:dkey-230i18.2 si:dkey-121n8.7 NLR family CARD domain containing 9 |
| chr3_-_4579214 | 0.04 |
ENSDART00000164456
|
ftr50
|
finTRIM family, member 50 |
| chr4_-_16883051 | 0.04 |
ENSDART00000124627
|
strap
|
serine/threonine kinase receptor associated protein |
| chr25_+_37297659 | 0.03 |
ENSDART00000086733
|
si:dkey-234i14.13
|
si:dkey-234i14.13 |
| chr2_+_42260021 | 0.03 |
ENSDART00000124702
ENSDART00000140203 ENSDART00000184079 ENSDART00000193349 |
ftr04
|
finTRIM family, member 4 |
| chr7_-_1918971 | 0.02 |
ENSDART00000181759
|
CABZ01040626.1
|
|
| chr6_-_58975010 | 0.02 |
ENSDART00000144911
ENSDART00000144514 |
mars
|
methionyl-tRNA synthetase |
| chr16_-_11961325 | 0.02 |
ENSDART00000143845
|
si:ch73-296e2.3
|
si:ch73-296e2.3 |
| chr15_-_34933560 | 0.02 |
ENSDART00000006288
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr8_+_3379815 | 0.02 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr22_-_9677264 | 0.02 |
ENSDART00000181737
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr2_-_1514001 | 0.02 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr20_+_46183505 | 0.02 |
ENSDART00000060799
|
taar13b
|
trace amine associated receptor 13b |
| chr15_-_5198191 | 0.02 |
ENSDART00000174064
|
or128-9
|
odorant receptor, family E, subfamily 128, member 9 |
| chr1_+_26467071 | 0.02 |
ENSDART00000112329
ENSDART00000159318 ENSDART00000193833 ENSDART00000011809 |
uso1
|
USO1 vesicle transport factor |
| chr2_-_40199780 | 0.01 |
ENSDART00000113901
|
ccl34a.4
|
chemokine (C-C motif) ligand 34a, duplicate 4 |
| chr15_-_5262942 | 0.01 |
ENSDART00000138003
|
or127-1
|
odorant receptor, family E, subfamily 127, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ptf1a+si:dkey-34f9.3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.4 | 1.9 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.3 | 1.7 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.2 | 1.0 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.2 | 1.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.2 | 2.2 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 0.6 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 2.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.5 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 1.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.2 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.3 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.2 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.2 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 1.1 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.0 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.0 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.7 | GO:0097610 | cleavage furrow(GO:0032154) recycling endosome membrane(GO:0055038) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.1 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.6 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.5 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.3 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.1 | 1.5 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.7 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 0.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 1.4 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.1 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 2.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.9 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |