Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
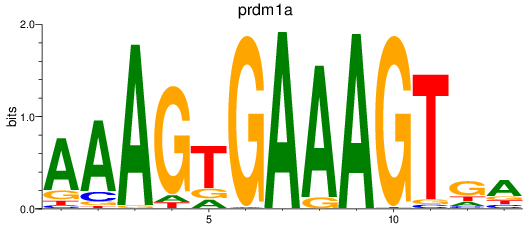
Results for prdm1a
Z-value: 0.56
Transcription factors associated with prdm1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prdm1a
|
ENSDARG00000002445 | PR domain containing 1a, with ZNF domain |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prdm1a | dr11_v1_chr16_-_7443388_7443388 | -0.74 | 2.3e-02 | Click! |
Activity profile of prdm1a motif
Sorted Z-values of prdm1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_10202718 | 0.63 |
ENSDART00000126668
|
pou6f2
|
POU class 6 homeobox 2 |
| chr5_-_17876709 | 0.50 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr10_-_45229877 | 0.47 |
ENSDART00000169281
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr24_+_26402110 | 0.46 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr5_-_26247671 | 0.45 |
ENSDART00000145187
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr4_-_17263210 | 0.42 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr5_-_26247973 | 0.41 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr13_-_24429066 | 0.41 |
ENSDART00000147514
ENSDART00000057588 |
cep68
|
centrosomal protein 68 |
| chr5_-_26247215 | 0.41 |
ENSDART00000136806
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr13_-_377256 | 0.40 |
ENSDART00000137398
|
ch1073-291c23.1
|
ch1073-291c23.1 |
| chr1_+_40034061 | 0.40 |
ENSDART00000011727
|
ccdc149b
|
coiled-coil domain containing 149b |
| chr2_-_7605121 | 0.39 |
ENSDART00000182099
|
CABZ01021599.1
|
|
| chr18_+_8812549 | 0.38 |
ENSDART00000017619
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr15_+_6459847 | 0.36 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr6_+_29861288 | 0.35 |
ENSDART00000166782
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr3_-_34586403 | 0.32 |
ENSDART00000151515
|
sept9a
|
septin 9a |
| chr17_+_5931530 | 0.31 |
ENSDART00000168326
ENSDART00000189790 |
znf513b
|
zinc finger protein 513b |
| chr13_-_21739142 | 0.30 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr16_-_50181206 | 0.30 |
ENSDART00000148478
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr18_-_12858016 | 0.30 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr23_-_30787932 | 0.30 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr10_-_45230081 | 0.30 |
ENSDART00000169845
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr15_+_40665310 | 0.29 |
ENSDART00000154187
ENSDART00000042082 |
fat3a
|
FAT atypical cadherin 3a |
| chr7_-_32782430 | 0.29 |
ENSDART00000173808
|
gas2b
|
growth arrest-specific 2b |
| chr16_-_25535430 | 0.29 |
ENSDART00000077420
|
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr8_-_19051906 | 0.28 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr12_+_33038757 | 0.28 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr23_+_43668756 | 0.28 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr12_-_4028079 | 0.27 |
ENSDART00000128676
|
si:ch211-180a12.2
|
si:ch211-180a12.2 |
| chr20_+_4793790 | 0.27 |
ENSDART00000153486
|
lgals8a
|
galectin 8a |
| chr7_+_27603211 | 0.27 |
ENSDART00000148782
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr3_+_823084 | 0.26 |
ENSDART00000153693
|
si:ch73-166o21.1
|
si:ch73-166o21.1 |
| chr1_-_55058795 | 0.26 |
ENSDART00000187293
|
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr13_+_7578111 | 0.25 |
ENSDART00000175431
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr10_+_38806719 | 0.25 |
ENSDART00000184416
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr19_+_8612839 | 0.25 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr10_-_31220558 | 0.24 |
ENSDART00000134866
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr5_-_57723929 | 0.24 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr20_+_4793516 | 0.24 |
ENSDART00000053875
|
lgals8a
|
galectin 8a |
| chr15_+_45591669 | 0.24 |
ENSDART00000157459
|
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr4_+_59061652 | 0.24 |
ENSDART00000150447
|
znf1127
|
zinc finger protein 1127 |
| chr2_-_16254782 | 0.23 |
ENSDART00000133708
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr3_+_16976095 | 0.23 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr2_+_30969029 | 0.22 |
ENSDART00000085242
|
lpin2
|
lipin 2 |
| chr17_+_1063988 | 0.22 |
ENSDART00000160400
|
gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr8_+_7737062 | 0.22 |
ENSDART00000166712
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr20_+_49787584 | 0.22 |
ENSDART00000193458
ENSDART00000181511 ENSDART00000185850 ENSDART00000185613 ENSDART00000191671 |
CABZ01078261.1
|
|
| chr18_-_16953978 | 0.21 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr5_-_60885935 | 0.21 |
ENSDART00000128350
|
rad51d
|
RAD51 paralog D |
| chr9_-_5045378 | 0.20 |
ENSDART00000149268
|
nr4a2a
|
nuclear receptor subfamily 4, group A, member 2a |
| chr22_+_1440702 | 0.20 |
ENSDART00000165677
|
si:dkeyp-53d3.3
|
si:dkeyp-53d3.3 |
| chr9_-_15208129 | 0.20 |
ENSDART00000137043
ENSDART00000131512 |
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr24_+_17260329 | 0.19 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr19_+_15485287 | 0.19 |
ENSDART00000182797
|
pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr16_-_9802998 | 0.19 |
ENSDART00000154217
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr21_+_20356458 | 0.19 |
ENSDART00000135816
|
pde6b
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr12_+_5189776 | 0.18 |
ENSDART00000081298
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr1_-_19599242 | 0.18 |
ENSDART00000088949
|
ECPAS
|
si:dkey-245p14.7 |
| chr15_+_7992906 | 0.18 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr25_-_30357027 | 0.18 |
ENSDART00000171137
|
pdia3
|
protein disulfide isomerase family A, member 3 |
| chr22_+_1683185 | 0.18 |
ENSDART00000158109
|
si:dkey-1b17.6
|
si:dkey-1b17.6 |
| chr14_+_35806605 | 0.18 |
ENSDART00000173093
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr8_-_14052349 | 0.18 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr9_+_46745348 | 0.18 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr15_-_25126842 | 0.17 |
ENSDART00000193670
|
vps53
|
vacuolar protein sorting 53 homolog (S. cerevisiae) |
| chr4_-_6373735 | 0.17 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr8_+_16004551 | 0.17 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr23_+_2825940 | 0.17 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr24_+_39027481 | 0.17 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr9_+_33158191 | 0.16 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr6_+_38896158 | 0.16 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr22_-_3564563 | 0.16 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr24_+_24285751 | 0.16 |
ENSDART00000122294
|
ythdf3
|
YTH N(6)-methyladenosine RNA binding protein 3 |
| chr3_-_34547000 | 0.16 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr3_+_938438 | 0.15 |
ENSDART00000165671
|
irgf4
|
immunity-related GTPase family, f4 |
| chr6_+_46697710 | 0.15 |
ENSDART00000154969
|
tespa1
|
thymocyte expressed, positive selection associated 1 |
| chr8_+_30112655 | 0.15 |
ENSDART00000099027
|
fancc
|
Fanconi anemia, complementation group C |
| chr3_-_31833266 | 0.15 |
ENSDART00000084932
ENSDART00000183617 |
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr4_-_20208166 | 0.15 |
ENSDART00000066891
|
gsap
|
gamma-secretase activating protein |
| chr19_+_15485556 | 0.15 |
ENSDART00000079014
|
pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr24_+_24923166 | 0.15 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr15_-_47929455 | 0.14 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr6_-_19271210 | 0.14 |
ENSDART00000163628
ENSDART00000159124 |
zgc:174863
|
zgc:174863 |
| chr17_-_44968407 | 0.14 |
ENSDART00000165850
|
ngb
|
neuroglobin |
| chr14_+_49233896 | 0.14 |
ENSDART00000159526
|
rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr3_+_47322494 | 0.14 |
ENSDART00000102202
|
ppap2d
|
phosphatidic acid phosphatase type 2D |
| chr3_+_12593558 | 0.14 |
ENSDART00000186891
ENSDART00000159252 |
abca3b
|
ATP-binding cassette, sub-family A (ABC1), member 3b |
| chr14_+_21159693 | 0.14 |
ENSDART00000192678
|
zgc:136929
|
zgc:136929 |
| chr23_-_33709964 | 0.13 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr1_-_55131439 | 0.13 |
ENSDART00000074083
|
rab1aa
|
RAB1A, member RAS oncogene family a |
| chr8_+_16004154 | 0.13 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr20_+_715739 | 0.13 |
ENSDART00000136768
|
myo6a
|
myosin VIa |
| chr13_+_11550454 | 0.13 |
ENSDART00000034935
ENSDART00000166908 |
desi2
|
desumoylating isopeptidase 2 |
| chr18_+_50275933 | 0.13 |
ENSDART00000143911
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr4_-_75641117 | 0.13 |
ENSDART00000182165
|
si:dkey-71l4.2
|
si:dkey-71l4.2 |
| chr1_+_48256950 | 0.13 |
ENSDART00000171182
|
SORCS3
|
sortilin related VPS10 domain containing receptor 3 |
| chr13_-_43637371 | 0.13 |
ENSDART00000127930
ENSDART00000084474 |
fam160b1
|
family with sequence similarity 160, member B1 |
| chr6_-_1587291 | 0.13 |
ENSDART00000067592
ENSDART00000178877 |
zgc:123305
|
zgc:123305 |
| chr1_+_31706668 | 0.13 |
ENSDART00000057879
|
as3mt
|
arsenite methyltransferase |
| chr23_-_3703569 | 0.13 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr23_-_21763598 | 0.12 |
ENSDART00000145408
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_+_10018466 | 0.12 |
ENSDART00000113551
|
trim2b
|
tripartite motif containing 2b |
| chr11_-_44485700 | 0.12 |
ENSDART00000186613
ENSDART00000172828 ENSDART00000113222 |
mfn1b
|
mitofusin 1b |
| chr22_-_10019768 | 0.12 |
ENSDART00000162077
|
BX324216.2
|
|
| chr5_-_64103863 | 0.12 |
ENSDART00000135014
ENSDART00000083684 |
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr1_+_44710955 | 0.12 |
ENSDART00000131296
ENSDART00000142187 |
ssrp1b
|
structure specific recognition protein 1b |
| chr10_+_28587446 | 0.12 |
ENSDART00000030138
ENSDART00000137508 |
cblb
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr5_+_32007627 | 0.11 |
ENSDART00000183061
|
scai
|
suppressor of cancer cell invasion |
| chr16_-_9802449 | 0.11 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr2_+_42005475 | 0.11 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr14_-_23684814 | 0.11 |
ENSDART00000024604
|
mars2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr18_+_50276337 | 0.11 |
ENSDART00000140352
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr7_-_8881514 | 0.11 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr21_+_40695345 | 0.10 |
ENSDART00000143594
|
ccdc82
|
coiled-coil domain containing 82 |
| chr21_+_5169154 | 0.10 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr18_+_14684115 | 0.10 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr7_+_11197940 | 0.10 |
ENSDART00000081346
|
cemip
|
cell migration inducing protein, hyaluronan binding |
| chr2_+_28995776 | 0.10 |
ENSDART00000138733
|
cdh12a
|
cadherin 12, type 2a (N-cadherin 2) |
| chr13_+_42011287 | 0.10 |
ENSDART00000131147
|
cyp1b1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr11_+_19433936 | 0.10 |
ENSDART00000162081
|
prickle2b
|
prickle homolog 2b |
| chr9_+_23765587 | 0.10 |
ENSDART00000145120
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr23_+_6232895 | 0.10 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr22_+_1508545 | 0.10 |
ENSDART00000164814
|
zgc:173486
|
zgc:173486 |
| chr1_-_8966046 | 0.10 |
ENSDART00000136198
|
socs1b
|
suppressor of cytokine signaling 1b |
| chr23_+_43870886 | 0.10 |
ENSDART00000102658
ENSDART00000149088 |
nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr5_-_42904329 | 0.10 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr4_-_74367912 | 0.09 |
ENSDART00000174199
ENSDART00000165257 |
ptprb
|
protein tyrosine phosphatase, receptor type, b |
| chr22_-_8509215 | 0.09 |
ENSDART00000140146
|
si:ch73-27e22.3
|
si:ch73-27e22.3 |
| chr14_-_12837052 | 0.09 |
ENSDART00000165004
ENSDART00000043180 |
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr18_+_50276653 | 0.09 |
ENSDART00000192120
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr18_-_44935174 | 0.09 |
ENSDART00000081025
|
pex16
|
peroxisomal biogenesis factor 16 |
| chr22_+_21153031 | 0.09 |
ENSDART00000166342
|
crlf1b
|
cytokine receptor-like factor 1b |
| chr11_-_44484952 | 0.09 |
ENSDART00000166674
ENSDART00000188016 |
mfn1b
|
mitofusin 1b |
| chr5_-_64213253 | 0.09 |
ENSDART00000171711
|
gpsm1a
|
G protein signaling modulator 1a |
| chr25_-_23745765 | 0.09 |
ENSDART00000089278
|
si:ch211-236l14.4
|
si:ch211-236l14.4 |
| chr7_+_37377335 | 0.08 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr7_+_5908077 | 0.08 |
ENSDART00000173404
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr12_-_16380593 | 0.08 |
ENSDART00000152133
|
hectd2
|
HECT domain containing 2 |
| chr1_+_51903413 | 0.08 |
ENSDART00000135861
|
nfixa
|
nuclear factor I/Xa |
| chr7_+_854237 | 0.08 |
ENSDART00000181029
|
zgc:175177
|
zgc:175177 |
| chr7_-_71384391 | 0.08 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr22_+_21152847 | 0.08 |
ENSDART00000007321
|
crlf1b
|
cytokine receptor-like factor 1b |
| chr23_-_6982966 | 0.08 |
ENSDART00000033774
ENSDART00000169965 |
edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr8_-_14080534 | 0.08 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr1_+_16574312 | 0.08 |
ENSDART00000187067
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr22_-_18164671 | 0.08 |
ENSDART00000014057
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr19_+_9032073 | 0.07 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr5_+_22406672 | 0.07 |
ENSDART00000141385
|
si:dkey-27p18.3
|
si:dkey-27p18.3 |
| chr6_-_58149165 | 0.07 |
ENSDART00000170752
|
tox2
|
TOX high mobility group box family member 2 |
| chr17_+_25955003 | 0.07 |
ENSDART00000156029
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr8_+_2878756 | 0.06 |
ENSDART00000168107
|
crb2b
|
crumbs family member 2b |
| chr3_-_367283 | 0.06 |
ENSDART00000155936
ENSDART00000161964 ENSDART00000158560 ENSDART00000135595 ENSDART00000145890 |
mhc1zaa
|
major histocompatibility complex class I ZAA |
| chr23_-_27479558 | 0.06 |
ENSDART00000013563
|
atf7a
|
activating transcription factor 7a |
| chr11_+_21910752 | 0.06 |
ENSDART00000114288
|
foxp4
|
forkhead box P4 |
| chr19_+_14115838 | 0.06 |
ENSDART00000192057
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr18_+_47313899 | 0.06 |
ENSDART00000192389
ENSDART00000189592 ENSDART00000184281 |
barx2
|
BARX homeobox 2 |
| chr4_-_9667380 | 0.06 |
ENSDART00000189671
ENSDART00000133214 |
dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr18_+_47313715 | 0.06 |
ENSDART00000138806
|
barx2
|
BARX homeobox 2 |
| chr22_-_23748284 | 0.06 |
ENSDART00000162005
|
cfhl2
|
complement factor H like 2 |
| chr11_-_38492269 | 0.05 |
ENSDART00000065613
|
elk4
|
ELK4, ETS-domain protein |
| chr20_-_4793450 | 0.05 |
ENSDART00000053870
|
galca
|
galactosylceramidase a |
| chr1_-_54938137 | 0.05 |
ENSDART00000147212
|
crtac1a
|
cartilage acidic protein 1a |
| chr21_-_7781555 | 0.05 |
ENSDART00000084380
ENSDART00000189131 |
aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr8_-_25690966 | 0.05 |
ENSDART00000033701
|
sema3ga
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ga |
| chr19_+_42716542 | 0.05 |
ENSDART00000144557
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr10_+_10210455 | 0.05 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr19_-_6774871 | 0.05 |
ENSDART00000170952
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr14_+_21754521 | 0.05 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr7_-_51627019 | 0.05 |
ENSDART00000174111
|
nhsl2
|
NHS-like 2 |
| chr2_+_42005217 | 0.05 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr22_-_18164835 | 0.05 |
ENSDART00000143189
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr8_-_24730185 | 0.05 |
ENSDART00000113197
|
prok1
|
prokineticin 1 |
| chr17_-_44968177 | 0.04 |
ENSDART00000075510
|
ngb
|
neuroglobin |
| chr5_-_64454459 | 0.04 |
ENSDART00000172321
ENSDART00000168030 |
brd3b
|
bromodomain containing 3b |
| chr12_+_5190049 | 0.04 |
ENSDART00000126667
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr22_-_34995333 | 0.04 |
ENSDART00000110900
|
kcnip2
|
Kv channel interacting protein 2 |
| chr11_-_38492564 | 0.04 |
ENSDART00000102850
|
elk4
|
ELK4, ETS-domain protein |
| chr5_-_30176349 | 0.04 |
ENSDART00000186584
|
adamts8a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8a |
| chr1_-_31083535 | 0.04 |
ENSDART00000138113
|
ppp1r9alb
|
protein phosphatase 1 regulatory subunit 9A-like B |
| chr24_-_20915050 | 0.04 |
ENSDART00000133763
|
si:ch211-161h7.5
|
si:ch211-161h7.5 |
| chr20_+_27096430 | 0.04 |
ENSDART00000153358
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr1_+_31942961 | 0.04 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr8_-_46457233 | 0.03 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr23_-_27123171 | 0.03 |
ENSDART00000023218
ENSDART00000146467 |
stat6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr10_+_9222476 | 0.03 |
ENSDART00000064973
ENSDART00000145879 |
paqr3b
|
progestin and adipoQ receptor family member IIIb |
| chr7_-_1992306 | 0.03 |
ENSDART00000053660
|
zgc:171424
|
zgc:171424 |
| chr22_-_23668356 | 0.03 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr21_+_30306369 | 0.03 |
ENSDART00000145050
ENSDART00000059420 |
lcp2a
|
lymphocyte cytosolic protein 2a |
| chr25_+_28693451 | 0.03 |
ENSDART00000148366
|
cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr25_+_10416583 | 0.03 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr2_-_24369087 | 0.03 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr20_-_6196989 | 0.03 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr3_-_42016693 | 0.03 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr1_-_21599219 | 0.03 |
ENSDART00000148327
|
adamtsl7
|
ADAMTS-like 7 |
| chr7_+_36552725 | 0.02 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr6_-_10780698 | 0.02 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr2_+_38608290 | 0.02 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr18_-_37252036 | 0.02 |
ENSDART00000132230
|
six5
|
SIX homeobox 5 |
| chr1_-_45636362 | 0.02 |
ENSDART00000181757
|
sept3
|
septin 3 |
| chr15_+_31886284 | 0.02 |
ENSDART00000156706
|
frya
|
furry homolog a (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of prdm1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.4 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.1 | 0.3 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.1 | 0.3 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.1 | 0.2 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.5 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.3 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.8 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.4 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.2 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.1 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.1 | GO:0061438 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 1.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |