Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
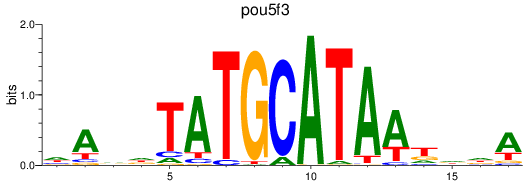
Results for pou5f3
Z-value: 0.52
Transcription factors associated with pou5f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou5f3
|
ENSDARG00000044774 | POU domain, class 5, transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou5f3 | dr11_v1_chr21_-_13690712_13690712 | 0.66 | 5.3e-02 | Click! |
Activity profile of pou5f3 motif
Sorted Z-values of pou5f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_41038141 | 1.47 |
ENSDART00000075620
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr8_+_41037541 | 1.28 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr1_-_55068941 | 1.06 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr5_-_30074332 | 1.06 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr7_+_33424044 | 0.99 |
ENSDART00000180260
|
glceb
|
glucuronic acid epimerase b |
| chr10_-_44560165 | 0.96 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr9_-_2945008 | 0.88 |
ENSDART00000183452
|
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr24_-_20641000 | 0.83 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr8_-_44238526 | 0.68 |
ENSDART00000061115
ENSDART00000132473 ENSDART00000138019 ENSDART00000133021 |
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr23_-_33709964 | 0.59 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr8_+_7740004 | 0.54 |
ENSDART00000170184
ENSDART00000187811 |
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr7_-_24644893 | 0.53 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr4_+_6869847 | 0.53 |
ENSDART00000036646
|
dock4b
|
dedicator of cytokinesis 4b |
| chr11_-_36001495 | 0.51 |
ENSDART00000190330
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr16_-_31351419 | 0.49 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr7_-_72155022 | 0.46 |
ENSDART00000187984
|
zmp:0000001168
|
zmp:0000001168 |
| chr17_+_28670132 | 0.40 |
ENSDART00000076344
ENSDART00000164981 ENSDART00000182851 |
hectd1
|
HECT domain containing 1 |
| chr24_+_7322116 | 0.36 |
ENSDART00000005804
|
XRCC2
|
X-ray repair cross complementing 2 |
| chr18_-_8030073 | 0.35 |
ENSDART00000151490
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr8_-_22698651 | 0.35 |
ENSDART00000181411
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr7_+_58843700 | 0.30 |
ENSDART00000159500
ENSDART00000158436 |
lypla1
|
lysophospholipase I |
| chr4_-_8903240 | 0.29 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr6_-_55399214 | 0.29 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr9_+_12444494 | 0.28 |
ENSDART00000102430
|
tmem41aa
|
transmembrane protein 41aa |
| chr18_+_20838786 | 0.23 |
ENSDART00000138692
|
ttc23
|
tetratricopeptide repeat domain 23 |
| chr2_-_20120904 | 0.16 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr25_+_26923193 | 0.13 |
ENSDART00000187364
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr13_+_31172833 | 0.10 |
ENSDART00000176378
|
CR931802.3
|
|
| chr5_+_29715040 | 0.09 |
ENSDART00000192563
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr20_-_34868814 | 0.08 |
ENSDART00000153049
|
stmn4
|
stathmin-like 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou5f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.8 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 1.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 1.1 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.7 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 1.0 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 1.0 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.2 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.3 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.5 | GO:0046847 | filopodium assembly(GO:0046847) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.7 | GO:0043186 | P granule(GO:0043186) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 1.0 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.1 | 1.1 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |