Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
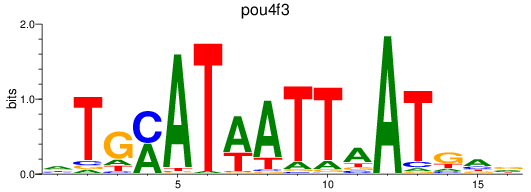
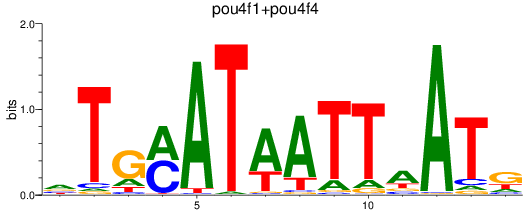
Results for pou4f3_pou4f1+pou4f4
Z-value: 0.94
Transcription factors associated with pou4f3_pou4f1+pou4f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou4f3
|
ENSDARG00000006206 | POU class 4 homeobox 3 |
|
pou4f1
|
ENSDARG00000005559 | POU class 4 homeobox 1 |
|
pou4f4
|
ENSDARG00000044375 | zgc |
|
pou4f4
|
ENSDARG00000111929 | zgc |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou4f3 | dr11_v1_chr9_+_54290896_54290896 | 0.93 | 3.4e-04 | Click! |
| pou4f1 | dr11_v1_chr6_+_4539953_4539953 | 0.75 | 2.1e-02 | Click! |
| zgc:158291 | dr11_v1_chr21_+_38312549_38312549 | 0.67 | 4.9e-02 | Click! |
Activity profile of pou4f3_pou4f1+pou4f4 motif
Sorted Z-values of pou4f3_pou4f1+pou4f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_21688176 | 1.32 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr21_+_26389391 | 1.21 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr24_-_3419998 | 1.07 |
ENSDART00000066839
|
slc35g2b
|
solute carrier family 35, member G2b |
| chr4_+_12111154 | 1.05 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr23_-_27345425 | 1.01 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr24_+_14713776 | 0.96 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr16_+_10318893 | 0.89 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr22_+_15507218 | 0.89 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr5_+_58372164 | 0.89 |
ENSDART00000057910
|
nrgna
|
neurogranin (protein kinase C substrate, RC3) a |
| chr20_+_34512130 | 0.89 |
ENSDART00000131637
|
prrx1b
|
paired related homeobox 1b |
| chr11_+_6819050 | 0.87 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr8_+_15254564 | 0.84 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr2_+_50608099 | 0.84 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr20_+_9355912 | 0.80 |
ENSDART00000142337
|
si:dkey-174m14.3
|
si:dkey-174m14.3 |
| chr9_-_22213297 | 0.80 |
ENSDART00000110656
ENSDART00000133149 |
crygm2d20
|
crystallin, gamma M2d20 |
| chr12_-_26406323 | 0.79 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr10_+_9550419 | 0.75 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr20_+_37661547 | 0.75 |
ENSDART00000007253
|
aig1
|
androgen-induced 1 (H. sapiens) |
| chr13_-_43599898 | 0.68 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr12_+_2446837 | 0.68 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr3_+_23731109 | 0.67 |
ENSDART00000131410
|
hoxb3a
|
homeobox B3a |
| chr5_-_30620625 | 0.65 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr16_+_10776688 | 0.63 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr13_-_32635859 | 0.62 |
ENSDART00000146249
ENSDART00000145395 ENSDART00000148040 ENSDART00000100650 |
matn3b
|
matrilin 3b |
| chr9_+_7030016 | 0.59 |
ENSDART00000148047
ENSDART00000148181 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr4_+_9669717 | 0.57 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr11_+_23957440 | 0.56 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr10_+_39084354 | 0.56 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr7_-_28148310 | 0.56 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr16_+_14029283 | 0.54 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr5_-_23429228 | 0.54 |
ENSDART00000049291
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr20_-_34801181 | 0.52 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr12_+_4260779 | 0.52 |
ENSDART00000081382
|
mmp25b
|
matrix metallopeptidase 25b |
| chr9_-_7683799 | 0.51 |
ENSDART00000102713
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr5_+_4366431 | 0.50 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr9_-_7684002 | 0.49 |
ENSDART00000016360
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr23_-_10177442 | 0.48 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr19_-_10196370 | 0.48 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr16_-_45398408 | 0.48 |
ENSDART00000004052
|
rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr22_+_5687615 | 0.47 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr23_+_21473103 | 0.46 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr8_+_10305400 | 0.45 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr2_+_55984788 | 0.44 |
ENSDART00000183599
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr20_+_22045089 | 0.43 |
ENSDART00000063564
ENSDART00000187013 ENSDART00000161552 ENSDART00000174478 ENSDART00000063568 ENSDART00000152247 |
nmu
|
neuromedin U |
| chr16_+_1353894 | 0.41 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr7_-_34225011 | 0.41 |
ENSDART00000049588
|
crybgx
|
crystallin beta gamma X |
| chr12_-_10705916 | 0.40 |
ENSDART00000164038
|
FO704786.1
|
|
| chr1_-_22861348 | 0.40 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr23_-_29376859 | 0.39 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr19_+_19772765 | 0.39 |
ENSDART00000182028
ENSDART00000161019 |
hoxa3a
|
homeobox A3a |
| chr3_-_28048475 | 0.38 |
ENSDART00000150888
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr10_+_38775408 | 0.37 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr14_-_18672561 | 0.36 |
ENSDART00000166730
ENSDART00000006998 |
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr19_-_47455944 | 0.35 |
ENSDART00000190005
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr19_+_30662529 | 0.34 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr18_-_23875370 | 0.34 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr25_+_24291156 | 0.33 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr10_-_22127942 | 0.33 |
ENSDART00000133374
|
ponzr2
|
plac8 onzin related protein 2 |
| chr20_+_20638034 | 0.32 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr13_+_23282549 | 0.32 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr3_+_26734162 | 0.32 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr9_-_33081978 | 0.32 |
ENSDART00000100918
|
zgc:172053
|
zgc:172053 |
| chr6_+_21740672 | 0.32 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr2_-_9818640 | 0.31 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr19_-_45960191 | 0.31 |
ENSDART00000052434
ENSDART00000172732 |
eif3hb
|
eukaryotic translation initiation factor 3, subunit H, b |
| chr10_+_38775959 | 0.30 |
ENSDART00000192990
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr13_+_31757331 | 0.29 |
ENSDART00000044282
|
hif1aa
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) a |
| chr6_-_35446110 | 0.29 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr8_+_668184 | 0.29 |
ENSDART00000183788
|
rnf165b
|
ring finger protein 165b |
| chr24_-_4765740 | 0.28 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr2_+_39108339 | 0.27 |
ENSDART00000085675
|
clstn2
|
calsyntenin 2 |
| chr25_+_3358701 | 0.26 |
ENSDART00000104877
|
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr22_-_23748284 | 0.26 |
ENSDART00000162005
|
cfhl2
|
complement factor H like 2 |
| chr11_+_23760470 | 0.26 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr25_+_19201231 | 0.25 |
ENSDART00000067323
|
hapln3
|
hyaluronan and proteoglycan link protein 3 |
| chr3_+_22578369 | 0.25 |
ENSDART00000187695
ENSDART00000182678 ENSDART00000112270 |
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr14_-_39031108 | 0.25 |
ENSDART00000026194
|
glra4a
|
glycine receptor, alpha 4a |
| chr6_-_41229787 | 0.25 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr23_+_23658474 | 0.25 |
ENSDART00000162838
|
agrn
|
agrin |
| chr13_-_31622195 | 0.24 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr20_-_10120442 | 0.24 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr2_+_22042745 | 0.24 |
ENSDART00000132039
|
tox
|
thymocyte selection-associated high mobility group box |
| chr23_+_21966447 | 0.24 |
ENSDART00000189378
|
lactbl1a
|
lactamase, beta-like 1a |
| chr18_-_47662696 | 0.23 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr24_+_3328354 | 0.23 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr19_+_37620342 | 0.23 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr11_+_37137196 | 0.23 |
ENSDART00000172950
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr6_+_38427570 | 0.23 |
ENSDART00000170612
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr15_-_21132480 | 0.23 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr7_-_71829649 | 0.23 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr13_+_27951688 | 0.23 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr16_-_46660680 | 0.23 |
ENSDART00000159209
ENSDART00000191929 |
tmem176l.4
|
transmembrane protein 176l.4 |
| chr13_+_12299997 | 0.22 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr19_-_43552252 | 0.22 |
ENSDART00000138308
|
gpr186
|
G protein-coupled receptor 186 |
| chr8_-_27858458 | 0.22 |
ENSDART00000132632
ENSDART00000136562 |
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr10_+_38708099 | 0.22 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr10_-_29892486 | 0.22 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr20_-_39103119 | 0.22 |
ENSDART00000143379
|
rcan2
|
regulator of calcineurin 2 |
| chr2_+_22702488 | 0.22 |
ENSDART00000076647
|
kif1ab
|
kinesin family member 1Ab |
| chr10_+_25726694 | 0.21 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr25_+_21833287 | 0.21 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr7_+_20518218 | 0.20 |
ENSDART00000052916
|
zgc:103586
|
zgc:103586 |
| chr22_+_16535575 | 0.20 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr17_+_33453689 | 0.20 |
ENSDART00000156894
|
rin3
|
Ras and Rab interactor 3 |
| chr23_+_11285662 | 0.19 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr17_-_15746898 | 0.19 |
ENSDART00000180079
|
cx52.7
|
connexin 52.7 |
| chr20_+_7180298 | 0.19 |
ENSDART00000143026
|
si:dkeyp-51f12.2
|
si:dkeyp-51f12.2 |
| chr16_-_9675982 | 0.19 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr13_-_39830999 | 0.19 |
ENSDART00000115089
|
zgc:171482
|
zgc:171482 |
| chr22_+_13886821 | 0.19 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr23_-_24195723 | 0.19 |
ENSDART00000145489
|
ano11
|
anoctamin 11 |
| chr15_+_45586471 | 0.19 |
ENSDART00000165699
|
cldn15lb
|
claudin 15-like b |
| chr2_+_25929619 | 0.19 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr15_-_23206562 | 0.19 |
ENSDART00000110360
|
ccdc153
|
coiled-coil domain containing 153 |
| chr9_-_33063083 | 0.18 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr15_-_21155641 | 0.18 |
ENSDART00000061098
ENSDART00000046443 |
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr14_+_8174828 | 0.18 |
ENSDART00000167228
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr23_+_21479958 | 0.18 |
ENSDART00000188302
ENSDART00000144320 |
si:dkey-1c11.1
|
si:dkey-1c11.1 |
| chr25_-_22187397 | 0.17 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr11_+_30057762 | 0.17 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr7_+_73397283 | 0.17 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr1_+_25783801 | 0.17 |
ENSDART00000102455
|
gucy1a1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr25_-_4148719 | 0.17 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr13_-_30700460 | 0.17 |
ENSDART00000139073
|
rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr12_+_8168272 | 0.17 |
ENSDART00000054092
|
arid5b
|
AT-rich interaction domain 5B |
| chr15_-_21239416 | 0.17 |
ENSDART00000155787
|
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr22_+_10651726 | 0.17 |
ENSDART00000145459
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr15_-_14552101 | 0.17 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr3_-_61336841 | 0.17 |
ENSDART00000155414
|
tecpr1b
|
tectonin beta-propeller repeat containing 1b |
| chr3_+_36313532 | 0.17 |
ENSDART00000151305
|
slc16a6b
|
solute carrier family 16, member 6b |
| chr9_-_40931637 | 0.17 |
ENSDART00000165103
|
C2orf88
|
Small membrane A-kinase anchor protein |
| chr6_+_8079974 | 0.17 |
ENSDART00000152071
|
si:ch211-207j7.2
|
si:ch211-207j7.2 |
| chr24_-_8831866 | 0.16 |
ENSDART00000066780
ENSDART00000143501 |
gcm2
|
glial cells missing homolog 2 (Drosophila) |
| chr5_+_39537195 | 0.16 |
ENSDART00000051256
|
fgf5
|
fibroblast growth factor 5 |
| chr20_-_11178022 | 0.16 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr1_-_21901589 | 0.16 |
ENSDART00000140553
|
frmpd1a
|
FERM and PDZ domain containing 1a |
| chr16_-_12953739 | 0.16 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr17_+_9009098 | 0.16 |
ENSDART00000180856
|
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr7_-_17690756 | 0.16 |
ENSDART00000173759
|
map4k2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr6_+_45494227 | 0.16 |
ENSDART00000159863
|
cntn4
|
contactin 4 |
| chr16_-_49505275 | 0.16 |
ENSDART00000160784
|
satb1b
|
SATB homeobox 1b |
| chr1_+_38758445 | 0.16 |
ENSDART00000136300
|
wdr17
|
WD repeat domain 17 |
| chr6_-_39489190 | 0.16 |
ENSDART00000151299
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr20_-_48172556 | 0.16 |
ENSDART00000097888
|
CABZ01059099.1
|
|
| chr21_+_5882300 | 0.16 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr23_-_24195519 | 0.15 |
ENSDART00000112370
ENSDART00000180377 |
ano11
|
anoctamin 11 |
| chr14_+_36231126 | 0.15 |
ENSDART00000141766
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr12_+_20618336 | 0.15 |
ENSDART00000084007
|
si:dkey-220f10.4
|
si:dkey-220f10.4 |
| chr23_+_6232895 | 0.15 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr1_-_23274393 | 0.15 |
ENSDART00000147800
ENSDART00000130277 ENSDART00000054340 ENSDART00000054338 |
rpl9
|
ribosomal protein L9 |
| chr3_-_23406964 | 0.15 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr18_+_15616167 | 0.15 |
ENSDART00000080454
|
slc17a8
|
solute carrier family 17 (vesicular glutamate transporter), member 8 |
| chr25_-_15040369 | 0.15 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr1_+_38758261 | 0.14 |
ENSDART00000182756
|
wdr17
|
WD repeat domain 17 |
| chr22_-_28226948 | 0.14 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr6_-_8295657 | 0.14 |
ENSDART00000185135
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr19_-_3574060 | 0.14 |
ENSDART00000105120
|
tmem170b
|
transmembrane protein 170B |
| chr5_-_54672763 | 0.14 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr6_+_13039951 | 0.13 |
ENSDART00000091700
|
catip
|
ciliogenesis associated TTC17 interacting protein |
| chr3_-_23407720 | 0.13 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr25_-_13614863 | 0.13 |
ENSDART00000121859
|
fa2h
|
fatty acid 2-hydroxylase |
| chr23_+_33907899 | 0.13 |
ENSDART00000159445
|
cs
|
citrate synthase |
| chr3_-_28665291 | 0.13 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr16_-_24605969 | 0.12 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr11_-_1956204 | 0.12 |
ENSDART00000185541
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr10_-_27566481 | 0.12 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr9_-_6661657 | 0.12 |
ENSDART00000133178
ENSDART00000113914 ENSDART00000061593 |
pou3f3a
|
POU class 3 homeobox 3a |
| chr12_+_20618522 | 0.12 |
ENSDART00000091452
|
si:dkey-220f10.4
|
si:dkey-220f10.4 |
| chr1_-_15797663 | 0.12 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr25_+_14507567 | 0.12 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr9_-_68934 | 0.12 |
ENSDART00000054594
ENSDART00000009389 |
il10rb
|
interleukin 10 receptor, beta |
| chr11_-_25777318 | 0.12 |
ENSDART00000079328
|
plch2b
|
phospholipase C, eta 2b |
| chr11_+_25101220 | 0.12 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr3_-_16055432 | 0.11 |
ENSDART00000123621
ENSDART00000023859 |
atp6v0ca
|
ATPase H+ transporting V0 subunit ca |
| chr1_+_31113951 | 0.11 |
ENSDART00000129362
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr18_-_40884087 | 0.11 |
ENSDART00000059194
|
snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr7_+_27317174 | 0.11 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr17_-_25649079 | 0.11 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr25_+_8407892 | 0.11 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr15_+_15403560 | 0.10 |
ENSDART00000049831
|
dhrs11b
|
dehydrogenase/reductase (SDR family) member 11b |
| chr9_-_25989989 | 0.10 |
ENSDART00000090052
|
arhgap15
|
Rho GTPase activating protein 15 |
| chr10_+_40214877 | 0.10 |
ENSDART00000109689
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr7_-_35515931 | 0.10 |
ENSDART00000193324
|
irx6a
|
iroquois homeobox 6a |
| chr17_-_37395460 | 0.10 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr11_+_44356707 | 0.10 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr9_+_44431174 | 0.10 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr14_-_36799280 | 0.09 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr6_+_39923052 | 0.09 |
ENSDART00000149019
|
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr19_+_33476557 | 0.09 |
ENSDART00000181800
|
triqk
|
triple QxxK/R motif containing |
| chr17_-_30975978 | 0.09 |
ENSDART00000051697
|
evla
|
Enah/Vasp-like a |
| chr23_-_30431333 | 0.08 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr22_-_17585618 | 0.08 |
ENSDART00000183123
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr20_-_40754794 | 0.08 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr14_+_30568961 | 0.08 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr17_+_2727807 | 0.08 |
ENSDART00000178759
|
kcnk10b
|
potassium channel, subfamily K, member 10b |
| chr20_-_39333657 | 0.08 |
ENSDART00000153720
ENSDART00000142164 |
ccl38.1
|
chemokine (C-C motif) ligand 38, duplicate 1 |
| chr7_-_25895189 | 0.08 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr15_+_32297441 | 0.08 |
ENSDART00000153657
|
trim3a
|
tripartite motif containing 3a |
| chr1_+_44491077 | 0.07 |
ENSDART00000073736
|
rtn4rl2a
|
reticulon 4 receptor-like 2 a |
| chr5_+_12888260 | 0.07 |
ENSDART00000175916
|
gal3st1a
|
galactose-3-O-sulfotransferase 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou4f3_pou4f1+pou4f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 0.9 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.3 | 1.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 0.6 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.5 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.3 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.1 | 0.2 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.2 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.2 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.2 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 1.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.2 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.8 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 1.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.1 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.6 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.4 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 0.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.5 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.2 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.1 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.0 | 0.2 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.3 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.0 | 0.4 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.2 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:2000651 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.1 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 1.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 1.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.5 | GO:1902749 | microtubule depolymerization(GO:0007019) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.3 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.1 | GO:0031101 | fin regeneration(GO:0031101) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.2 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.8 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.5 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.5 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |