Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
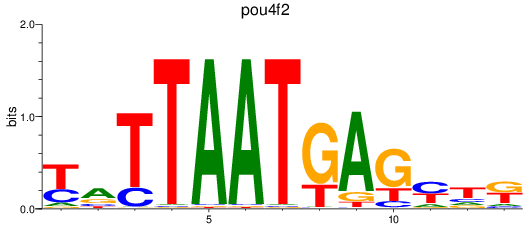
Results for pou4f2
Z-value: 0.43
Transcription factors associated with pou4f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou4f2
|
ENSDARG00000069737 | POU class 4 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou4f2 | dr11_v1_chr1_+_36437585_36437585 | 0.16 | 6.7e-01 | Click! |
Activity profile of pou4f2 motif
Sorted Z-values of pou4f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_36939749 | 0.43 |
ENSDART00000005382
|
gadd45bb
|
growth arrest and DNA-damage-inducible, beta b |
| chr20_-_6532462 | 0.37 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr13_+_1089942 | 0.25 |
ENSDART00000054322
|
cnrip1b
|
cannabinoid receptor interacting protein 1b |
| chr9_+_50001746 | 0.22 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr9_-_8314028 | 0.18 |
ENSDART00000102739
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr5_-_26247973 | 0.17 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr2_-_21082695 | 0.17 |
ENSDART00000032502
|
nebl
|
nebulette |
| chr19_-_18313303 | 0.17 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr10_+_5268054 | 0.16 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr17_+_26965351 | 0.15 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr21_+_20396858 | 0.15 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr12_-_35787801 | 0.14 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr22_+_10698549 | 0.14 |
ENSDART00000081228
|
abhd14a
|
abhydrolase domain containing 14A |
| chr19_-_31402429 | 0.14 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr21_+_76739 | 0.14 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr9_+_21793565 | 0.13 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr7_+_49862837 | 0.12 |
ENSDART00000174315
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr14_-_7409364 | 0.12 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr17_-_29902187 | 0.11 |
ENSDART00000009104
|
esrrb
|
estrogen-related receptor beta |
| chr13_-_14487524 | 0.11 |
ENSDART00000141103
|
gfra4a
|
GDNF family receptor alpha 4a |
| chr13_+_31180084 | 0.11 |
ENSDART00000133774
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr5_+_44064764 | 0.11 |
ENSDART00000143843
|
si:dkey-84j12.1
|
si:dkey-84j12.1 |
| chr13_-_33194647 | 0.11 |
ENSDART00000134383
|
trip11
|
thyroid hormone receptor interactor 11 |
| chr9_-_29003245 | 0.11 |
ENSDART00000183391
ENSDART00000188836 |
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr20_+_41756996 | 0.10 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr17_+_51499789 | 0.10 |
ENSDART00000187701
|
CABZ01067581.1
|
|
| chr2_+_23006792 | 0.10 |
ENSDART00000027782
|
mknk2a
|
MAP kinase interacting serine/threonine kinase 2a |
| chr14_-_31465905 | 0.10 |
ENSDART00000173108
|
gpc3
|
glypican 3 |
| chr1_-_22512063 | 0.10 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr10_+_40284003 | 0.10 |
ENSDART00000062795
ENSDART00000193825 ENSDART00000113582 |
git2b
|
G protein-coupled receptor kinase interacting ArfGAP 2b |
| chr14_+_25817628 | 0.09 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr9_-_43538328 | 0.09 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr23_-_28141419 | 0.09 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr24_+_19593197 | 0.09 |
ENSDART00000151923
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr9_-_3934963 | 0.09 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr11_-_31226578 | 0.09 |
ENSDART00000109698
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr16_+_25011994 | 0.09 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr15_+_45994123 | 0.08 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr11_+_44502410 | 0.08 |
ENSDART00000172998
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr2_+_29249561 | 0.08 |
ENSDART00000099157
|
cdh18a
|
cadherin 18, type 2a |
| chr3_-_30434016 | 0.08 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr20_+_38458084 | 0.08 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr14_-_7306983 | 0.08 |
ENSDART00000158914
|
si:ch211-51f19.1
|
si:ch211-51f19.1 |
| chr3_-_30384353 | 0.08 |
ENSDART00000186832
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr8_-_40555340 | 0.08 |
ENSDART00000163348
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr5_-_68782641 | 0.08 |
ENSDART00000141699
|
mepce
|
methylphosphate capping enzyme |
| chr15_-_5720583 | 0.07 |
ENSDART00000158034
ENSDART00000190332 ENSDART00000109599 |
urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr2_+_12302527 | 0.07 |
ENSDART00000091298
ENSDART00000182569 |
arhgap21b
|
Rho GTPase activating protein 21b |
| chr11_-_3860534 | 0.07 |
ENSDART00000082425
|
gata2a
|
GATA binding protein 2a |
| chr16_+_40043673 | 0.07 |
ENSDART00000102552
ENSDART00000125484 |
trmt11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr16_-_12173399 | 0.07 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr2_-_30784198 | 0.07 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr7_-_7764287 | 0.07 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr20_-_31743553 | 0.07 |
ENSDART00000087405
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr7_-_69185124 | 0.07 |
ENSDART00000182217
ENSDART00000191359 |
usp10
|
ubiquitin specific peptidase 10 |
| chr10_-_35103208 | 0.07 |
ENSDART00000192734
|
zgc:110006
|
zgc:110006 |
| chr12_+_32729470 | 0.07 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr17_-_37395460 | 0.07 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr9_-_32158288 | 0.07 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr19_+_42470396 | 0.07 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr20_+_13781779 | 0.07 |
ENSDART00000142999
ENSDART00000152471 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr8_+_52314542 | 0.07 |
ENSDART00000013059
ENSDART00000125241 |
dbnlb
|
drebrin-like b |
| chr17_-_49438873 | 0.07 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr7_+_53498152 | 0.07 |
ENSDART00000184497
|
znf609b
|
zinc finger protein 609b |
| chr21_+_25226558 | 0.07 |
ENSDART00000168480
|
sycn.2
|
syncollin, tandem duplicate 2 |
| chr4_+_9836465 | 0.07 |
ENSDART00000004879
|
hsp90b1
|
heat shock protein 90, beta (grp94), member 1 |
| chr9_-_50001606 | 0.06 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr5_+_37966505 | 0.06 |
ENSDART00000127648
|
pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr18_-_2433011 | 0.06 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr11_+_3585934 | 0.06 |
ENSDART00000055694
|
cdab
|
cytidine deaminase b |
| chr17_+_8183393 | 0.06 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr8_-_4327473 | 0.06 |
ENSDART00000134378
|
cux2b
|
cut-like homeobox 2b |
| chr11_-_3860199 | 0.06 |
ENSDART00000082420
|
gata2a
|
GATA binding protein 2a |
| chr10_+_2582254 | 0.06 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin like 2 |
| chr10_+_38775959 | 0.06 |
ENSDART00000192990
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr4_-_2637689 | 0.06 |
ENSDART00000192550
ENSDART00000021953 ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
| chr11_-_2478374 | 0.06 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr23_+_41912151 | 0.06 |
ENSDART00000191115
|
podn
|
podocan |
| chr19_+_1688727 | 0.05 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr10_+_38775408 | 0.05 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr2_+_50608099 | 0.05 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr2_+_16696052 | 0.05 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr16_+_25107344 | 0.05 |
ENSDART00000033211
|
zgc:66448
|
zgc:66448 |
| chr19_-_5103313 | 0.05 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr25_+_5972690 | 0.05 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr8_+_12951155 | 0.05 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr25_-_13842618 | 0.05 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr18_-_43993614 | 0.05 |
ENSDART00000148535
ENSDART00000077357 |
bcl9l
|
B cell CLL/lymphoma 9-like |
| chr5_+_51443009 | 0.05 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr20_+_18325556 | 0.04 |
ENSDART00000123559
|
znf521
|
zinc finger protein 521 |
| chr14_+_28486213 | 0.04 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr17_+_6765621 | 0.04 |
ENSDART00000156637
ENSDART00000007622 |
afg1la
|
AFG1 like ATPase a |
| chr25_+_28555584 | 0.04 |
ENSDART00000157046
|
SLC15A5
|
si:ch211-190o6.3 |
| chr6_-_46735380 | 0.04 |
ENSDART00000103455
|
tarbp2
|
TAR (HIV) RNA binding protein 2 |
| chr20_-_24182689 | 0.04 |
ENSDART00000171184
|
map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr1_-_53756851 | 0.04 |
ENSDART00000122445
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr17_+_11675362 | 0.04 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr24_+_24808955 | 0.04 |
ENSDART00000080963
|
dnajc5b
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr6_-_6248893 | 0.04 |
ENSDART00000124662
|
rtn4a
|
reticulon 4a |
| chr17_+_26352372 | 0.04 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr21_+_36623162 | 0.04 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr2_+_29249204 | 0.04 |
ENSDART00000168957
|
cdh18a
|
cadherin 18, type 2a |
| chr21_-_25612658 | 0.04 |
ENSDART00000115276
|
fibpb
|
fibroblast growth factor (acidic) intracellular binding protein b |
| chr11_-_37509001 | 0.04 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr3_-_61362398 | 0.03 |
ENSDART00000156177
|
si:dkey-111k8.3
|
si:dkey-111k8.3 |
| chr16_-_26074529 | 0.03 |
ENSDART00000148653
ENSDART00000148923 |
tmem145
|
transmembrane protein 145 |
| chr10_+_38526496 | 0.03 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr16_-_11859309 | 0.03 |
ENSDART00000145754
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr2_-_3045861 | 0.03 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr13_-_42400647 | 0.03 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr1_+_144284 | 0.03 |
ENSDART00000064061
|
prozb
|
protein Z, vitamin K-dependent plasma glycoprotein b |
| chr13_-_16066997 | 0.03 |
ENSDART00000184790
|
SPATA48
|
spermatogenesis associated 48 |
| chr18_-_1185772 | 0.03 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr15_+_31332552 | 0.03 |
ENSDART00000134933
ENSDART00000173915 |
or119-2
|
odorant receptor, family F, subfamily 119, member 2 |
| chr7_-_17779644 | 0.03 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr4_+_11695979 | 0.03 |
ENSDART00000137736
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr9_+_29430432 | 0.03 |
ENSDART00000125632
|
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr6_-_35046735 | 0.03 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr10_+_2899108 | 0.02 |
ENSDART00000147031
|
erap1a
|
endoplasmic reticulum aminopeptidase 1a |
| chr14_+_29941445 | 0.02 |
ENSDART00000181761
|
fam149a
|
family with sequence similarity 149 member A |
| chr6_-_12172424 | 0.02 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr15_+_43398317 | 0.02 |
ENSDART00000182528
ENSDART00000172154 ENSDART00000187688 |
actn4
|
actinin, alpha 4 |
| chr5_-_38155005 | 0.02 |
ENSDART00000097770
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr6_+_27146671 | 0.02 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr20_-_18736281 | 0.02 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr1_+_25801648 | 0.02 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr3_-_1970820 | 0.02 |
ENSDART00000136467
|
si:ch211-254c8.3
|
si:ch211-254c8.3 |
| chr17_+_28706946 | 0.02 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr1_+_10318089 | 0.02 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr8_+_44714336 | 0.02 |
ENSDART00000145801
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr16_+_11242443 | 0.02 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr3_-_33574576 | 0.02 |
ENSDART00000184881
|
CR847537.1
|
|
| chr13_+_36622100 | 0.02 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr14_-_32959851 | 0.02 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr19_-_5103141 | 0.02 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr11_-_42918971 | 0.01 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr20_-_37813863 | 0.01 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr3_-_32169754 | 0.01 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr16_-_28091597 | 0.01 |
ENSDART00000166595
|
CR626941.1
|
|
| chr8_-_13454281 | 0.01 |
ENSDART00000141959
|
CR354547.2
|
|
| chr15_+_14826966 | 0.01 |
ENSDART00000157912
ENSDART00000171133 |
or130-1
|
odorant receptor, family H, subfamily 130, member 1 |
| chr9_+_38088331 | 0.01 |
ENSDART00000123749
|
cacnb4a
|
calcium channel, voltage-dependent, beta 4a subunit |
| chr18_+_2228737 | 0.01 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr16_-_12173554 | 0.01 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr7_-_18416741 | 0.01 |
ENSDART00000097882
|
sstr1b
|
somatostatin receptor 1b |
| chr12_+_17154655 | 0.01 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr4_-_28335184 | 0.01 |
ENSDART00000178149
ENSDART00000043737 |
celsr1a
|
cadherin EGF LAG seven-pass G-type receptor 1a |
| chr7_+_25059845 | 0.01 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr21_+_43328685 | 0.01 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr9_-_43082945 | 0.00 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr14_+_46287296 | 0.00 |
ENSDART00000183620
|
cabp2b
|
calcium binding protein 2b |
| chr15_+_5280858 | 0.00 |
ENSDART00000174242
|
or121-1
|
odorant receptor, family E, subfamily 121, member 1 |
| chr13_+_31177934 | 0.00 |
ENSDART00000144568
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr16_-_40043322 | 0.00 |
ENSDART00000145278
|
si:dkey-29b11.3
|
si:dkey-29b11.3 |
| chr22_+_2403068 | 0.00 |
ENSDART00000132925
ENSDART00000132569 |
zgc:112977
|
zgc:112977 |
| chr6_-_40466861 | 0.00 |
ENSDART00000062724
|
crbn
|
cereblon |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou4f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.1 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.0 | GO:0070920 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.1 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.1 | GO:0035889 | otolith tethering(GO:0035889) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.0 | GO:0070578 | RISC-loading complex(GO:0070578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.0 | GO:0070883 | pre-miRNA binding(GO:0070883) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |