Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
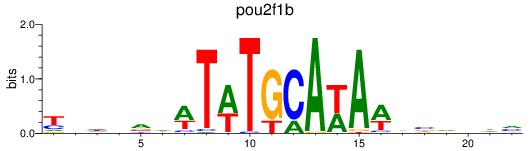
Results for pou2f1b
Z-value: 0.76
Transcription factors associated with pou2f1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f1b
|
ENSDARG00000007996 | POU class 2 homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f1b | dr11_v1_chr9_+_34331368_34331368 | 0.51 | 1.6e-01 | Click! |
Activity profile of pou2f1b motif
Sorted Z-values of pou2f1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_6618574 | 2.53 |
ENSDART00000184486
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr20_-_41992878 | 2.04 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr21_+_1119046 | 1.74 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr17_-_6641535 | 1.53 |
ENSDART00000154540
ENSDART00000180384 |
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr19_+_2835240 | 1.33 |
ENSDART00000190838
|
CDCP1
|
CUB domain containing protein 1 |
| chr19_-_27570333 | 1.22 |
ENSDART00000146562
ENSDART00000179060 |
si:dkeyp-46h3.5
si:dkeyp-46h3.8
|
si:dkeyp-46h3.5 si:dkeyp-46h3.8 |
| chr15_+_38308421 | 1.16 |
ENSDART00000129941
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr12_+_22560067 | 1.12 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr22_+_14836040 | 1.08 |
ENSDART00000180951
|
gtpbp1l
|
GTP binding protein 1, like |
| chr14_-_46198373 | 1.04 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr2_+_37430917 | 1.03 |
ENSDART00000140558
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr1_-_55262763 | 1.00 |
ENSDART00000152769
|
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr5_-_11809710 | 0.91 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr6_+_28208973 | 0.90 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr24_-_21150100 | 0.89 |
ENSDART00000154249
ENSDART00000193256 |
gramd1c
|
GRAM domain containing 1c |
| chr13_+_33462232 | 0.86 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr14_-_45967981 | 0.85 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr20_-_49889111 | 0.85 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr19_-_27550768 | 0.83 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr15_-_18209672 | 0.81 |
ENSDART00000141508
ENSDART00000136280 |
btr16
|
bloodthirsty-related gene family, member 16 |
| chr16_+_50434668 | 0.79 |
ENSDART00000193500
|
IGLON5
|
zgc:110372 |
| chr17_-_25831569 | 0.73 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr18_+_3579829 | 0.73 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr23_+_12160900 | 0.72 |
ENSDART00000136046
|
ppp1r3da
|
protein phosphatase 1, regulatory subunit 3Da |
| chr22_+_24318131 | 0.72 |
ENSDART00000187360
ENSDART00000165618 |
ccdc50
|
coiled-coil domain containing 50 |
| chr5_-_68779747 | 0.72 |
ENSDART00000192636
ENSDART00000188039 |
mepce
|
methylphosphate capping enzyme |
| chr10_+_34001444 | 0.71 |
ENSDART00000149934
|
kl
|
klotho |
| chr7_+_19600262 | 0.71 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr8_-_44238526 | 0.68 |
ENSDART00000061115
ENSDART00000132473 ENSDART00000138019 ENSDART00000133021 |
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr12_-_35582683 | 0.67 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr14_-_7409364 | 0.67 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr22_+_14836291 | 0.65 |
ENSDART00000122740
|
gtpbp1l
|
GTP binding protein 1, like |
| chr12_-_22238004 | 0.65 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr15_+_5132439 | 0.64 |
ENSDART00000010350
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr2_+_6253246 | 0.64 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr18_+_8917766 | 0.63 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr15_+_29393519 | 0.62 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr14_-_45967712 | 0.61 |
ENSDART00000043751
ENSDART00000141357 |
macrod1
|
MACRO domain containing 1 |
| chr19_+_1510971 | 0.61 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr19_+_1688727 | 0.61 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr6_+_33931095 | 0.60 |
ENSDART00000191909
|
orc1
|
origin recognition complex, subunit 1 |
| chr3_-_36153591 | 0.60 |
ENSDART00000190515
|
engase
|
endo-beta-N-acetylglucosaminidase |
| chr16_-_42770064 | 0.59 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr24_-_12770357 | 0.59 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr10_-_244745 | 0.59 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr18_+_22287084 | 0.59 |
ENSDART00000151919
ENSDART00000181644 |
ctcf
|
CCCTC-binding factor (zinc finger protein) |
| chr5_-_13206878 | 0.58 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr8_-_39884359 | 0.57 |
ENSDART00000131372
|
mlec
|
malectin |
| chr2_-_21438492 | 0.57 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr22_-_37834312 | 0.56 |
ENSDART00000076128
|
ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr4_-_72643171 | 0.55 |
ENSDART00000130126
|
CABZ01054394.1
|
|
| chr24_+_39586701 | 0.54 |
ENSDART00000136447
|
si:dkey-161j23.7
|
si:dkey-161j23.7 |
| chr10_-_41157135 | 0.54 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr5_-_65662996 | 0.54 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr9_-_8297344 | 0.54 |
ENSDART00000180945
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr16_+_53590586 | 0.53 |
ENSDART00000083534
|
gpatch3
|
G patch domain containing 3 |
| chr24_+_7322116 | 0.53 |
ENSDART00000005804
|
XRCC2
|
X-ray repair cross complementing 2 |
| chr23_+_27789795 | 0.53 |
ENSDART00000141458
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr11_+_30647545 | 0.53 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr21_-_39546737 | 0.53 |
ENSDART00000006971
|
sept4a
|
septin 4a |
| chr14_-_16754262 | 0.52 |
ENSDART00000001159
|
mgat4b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr6_+_54687495 | 0.52 |
ENSDART00000189514
|
smpd2b
|
sphingomyelin phosphodiesterase 2b, neutral membrane (neutral sphingomyelinase) |
| chr8_+_41038141 | 0.51 |
ENSDART00000075620
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr25_-_9013963 | 0.51 |
ENSDART00000073912
|
rag2
|
recombination activating gene 2 |
| chr9_-_30502010 | 0.51 |
ENSDART00000149483
|
si:dkey-229b18.3
|
si:dkey-229b18.3 |
| chr21_-_3613702 | 0.51 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr2_-_9915814 | 0.50 |
ENSDART00000091644
ENSDART00000177556 |
abi1b
|
abl-interactor 1b |
| chr15_-_30815826 | 0.49 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr5_-_1487256 | 0.49 |
ENSDART00000149599
ENSDART00000148411 ENSDART00000092087 ENSDART00000148464 |
golga2
|
golgin A2 |
| chr21_+_45502621 | 0.49 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr3_+_7771420 | 0.49 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr4_+_6869847 | 0.48 |
ENSDART00000036646
|
dock4b
|
dedicator of cytokinesis 4b |
| chr6_-_17999776 | 0.48 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr17_+_28628404 | 0.48 |
ENSDART00000032975
ENSDART00000143607 |
heatr5a
|
HEAT repeat containing 5a |
| chr10_+_23060391 | 0.48 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr23_+_44665230 | 0.48 |
ENSDART00000144958
|
camta2
|
calmodulin binding transcription activator 2 |
| chr21_+_76739 | 0.48 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr1_-_51038885 | 0.48 |
ENSDART00000035150
|
spast
|
spastin |
| chr16_-_43011470 | 0.47 |
ENSDART00000131898
ENSDART00000142003 ENSDART00000017966 |
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr25_+_27937585 | 0.47 |
ENSDART00000151994
|
cadps2
|
Ca++-dependent secretion activator 2 |
| chr11_-_21404044 | 0.47 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr3_-_59981476 | 0.47 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr20_+_4221978 | 0.47 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr9_-_12659140 | 0.47 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr21_-_43398457 | 0.47 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr21_-_34926619 | 0.46 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr25_+_27937789 | 0.46 |
ENSDART00000023890
|
cadps2
|
Ca++-dependent secretion activator 2 |
| chr25_+_22320738 | 0.45 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr9_+_48755638 | 0.45 |
ENSDART00000047401
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr22_+_14051894 | 0.44 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr19_-_977849 | 0.44 |
ENSDART00000172303
|
CABZ01088282.1
|
|
| chr19_+_43684376 | 0.44 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr17_+_20576848 | 0.44 |
ENSDART00000183203
|
SAMD8
|
zgc:162183 |
| chr17_+_1544903 | 0.44 |
ENSDART00000156244
ENSDART00000112183 |
cep170b
|
centrosomal protein 170B |
| chr8_+_41037541 | 0.44 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr7_-_72155022 | 0.44 |
ENSDART00000187984
|
zmp:0000001168
|
zmp:0000001168 |
| chr5_-_8711157 | 0.44 |
ENSDART00000062957
|
paip1
|
poly(A) binding protein interacting protein 1 |
| chr6_-_16394528 | 0.44 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr4_-_3353595 | 0.44 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr18_-_15551360 | 0.44 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr20_-_34754617 | 0.43 |
ENSDART00000148066
|
znf395b
|
zinc finger protein 395b |
| chr15_-_14038227 | 0.43 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr2_-_21625493 | 0.43 |
ENSDART00000137169
|
zgc:55781
|
zgc:55781 |
| chr10_+_29771256 | 0.43 |
ENSDART00000193195
|
hyou1
|
hypoxia up-regulated 1 |
| chr7_+_13582256 | 0.42 |
ENSDART00000158477
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr20_-_18794789 | 0.42 |
ENSDART00000003834
|
ccm2
|
cerebral cavernous malformation 2 |
| chr11_-_26576754 | 0.42 |
ENSDART00000191733
ENSDART00000002846 |
st3gal8
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 8 |
| chr25_+_34862225 | 0.41 |
ENSDART00000149782
|
CHST6
|
zgc:194879 |
| chr9_+_17983463 | 0.41 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr7_-_19364813 | 0.41 |
ENSDART00000173977
|
ntn4
|
netrin 4 |
| chr11_-_38083397 | 0.41 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr12_-_4243268 | 0.40 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr7_-_26497947 | 0.40 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr18_-_26715655 | 0.40 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr11_-_21404358 | 0.40 |
ENSDART00000129062
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr24_-_41180149 | 0.40 |
ENSDART00000019975
|
acvr2ba
|
activin A receptor type 2Ba |
| chr13_-_43637371 | 0.40 |
ENSDART00000127930
ENSDART00000084474 |
fam160b1
|
family with sequence similarity 160, member B1 |
| chr7_-_45852270 | 0.40 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr12_-_35582521 | 0.40 |
ENSDART00000162175
ENSDART00000168958 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr11_+_5817202 | 0.40 |
ENSDART00000126084
|
ctdspl3
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 3 |
| chr20_-_9123052 | 0.40 |
ENSDART00000125133
|
mysm1
|
Myb-like, SWIRM and MPN domains 1 |
| chr11_-_18791834 | 0.40 |
ENSDART00000156431
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr14_+_16083818 | 0.39 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr24_+_39135419 | 0.39 |
ENSDART00000180941
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr12_-_6551681 | 0.38 |
ENSDART00000145413
|
si:ch211-253p2.2
|
si:ch211-253p2.2 |
| chr17_+_28102487 | 0.38 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr8_-_47329755 | 0.38 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr3_-_21106093 | 0.38 |
ENSDART00000156566
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr13_+_11550454 | 0.38 |
ENSDART00000034935
ENSDART00000166908 |
desi2
|
desumoylating isopeptidase 2 |
| chr2_+_44615000 | 0.38 |
ENSDART00000188826
ENSDART00000113232 |
yeats2
|
YEATS domain containing 2 |
| chr13_-_23756700 | 0.38 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr21_-_11054605 | 0.38 |
ENSDART00000191378
ENSDART00000084061 |
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr23_-_33775145 | 0.37 |
ENSDART00000132147
ENSDART00000027959 ENSDART00000160116 |
racgap1
|
Rac GTPase activating protein 1 |
| chr20_-_9123296 | 0.37 |
ENSDART00000188495
|
mysm1
|
Myb-like, SWIRM and MPN domains 1 |
| chr18_-_26715156 | 0.37 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr19_+_15485287 | 0.37 |
ENSDART00000182797
|
pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr9_-_12443726 | 0.37 |
ENSDART00000102434
|
ehhadh
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr14_-_4044545 | 0.37 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr6_+_30491129 | 0.37 |
ENSDART00000088142
|
wwc3
|
WWC family member 3 |
| chr23_-_36857964 | 0.37 |
ENSDART00000188822
ENSDART00000134061 ENSDART00000093061 |
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr10_+_31809226 | 0.37 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr21_+_36623162 | 0.37 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr21_+_45502773 | 0.36 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr1_+_19434198 | 0.36 |
ENSDART00000012552
|
clockb
|
clock circadian regulator b |
| chr24_-_20641000 | 0.36 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr10_+_399363 | 0.36 |
ENSDART00000147449
|
si:ch211-242f23.8
|
si:ch211-242f23.8 |
| chr16_+_52847079 | 0.36 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr3_-_59981162 | 0.36 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr20_-_14941046 | 0.36 |
ENSDART00000152775
ENSDART00000152516 ENSDART00000021394 |
mettl13
|
methyltransferase like 13 |
| chr25_+_25516016 | 0.35 |
ENSDART00000188980
|
phrf1
|
PHD and ring finger domains 1 |
| chr5_+_31350927 | 0.35 |
ENSDART00000028237
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr15_+_34592215 | 0.35 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr21_+_30721733 | 0.35 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr21_-_35324091 | 0.35 |
ENSDART00000185042
|
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr21_-_11367271 | 0.35 |
ENSDART00000151000
ENSDART00000151465 |
zgc:162472
|
zgc:162472 |
| chr21_-_11054876 | 0.35 |
ENSDART00000146576
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr20_-_39789036 | 0.35 |
ENSDART00000086405
ENSDART00000098253 |
rnf217
|
ring finger protein 217 |
| chr4_+_5341592 | 0.35 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr23_+_16807209 | 0.35 |
ENSDART00000141966
|
zgc:114081
|
zgc:114081 |
| chr8_-_5850472 | 0.35 |
ENSDART00000179217
|
CABZ01102147.1
|
|
| chr12_+_35011899 | 0.34 |
ENSDART00000153007
ENSDART00000153020 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr5_+_71924175 | 0.34 |
ENSDART00000115182
ENSDART00000170215 |
nup214
|
nucleoporin 214 |
| chr4_+_5832311 | 0.34 |
ENSDART00000121743
ENSDART00000158233 ENSDART00000165187 |
si:ch73-352p4.5
|
si:ch73-352p4.5 |
| chr12_-_16435150 | 0.34 |
ENSDART00000086211
|
pcgf5b
|
polycomb group ring finger 5b |
| chr11_+_25596038 | 0.34 |
ENSDART00000140856
|
ccdc120
|
coiled-coil domain containing 120 |
| chr1_+_32051581 | 0.34 |
ENSDART00000146602
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr19_-_30800004 | 0.34 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr19_+_42432625 | 0.34 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr16_+_26735894 | 0.33 |
ENSDART00000191605
ENSDART00000185175 |
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr14_-_20891745 | 0.33 |
ENSDART00000106205
|
ids
|
iduronate 2-sulfatase |
| chr3_+_29476085 | 0.33 |
ENSDART00000184495
ENSDART00000181058 |
fam83fa
|
family with sequence similarity 83, member Fa |
| chr1_+_51615672 | 0.33 |
ENSDART00000165117
|
zgc:165656
|
zgc:165656 |
| chr24_-_20321012 | 0.33 |
ENSDART00000163683
|
map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr25_+_34407529 | 0.32 |
ENSDART00000156751
|
OTUD7A
|
si:dkey-37f18.2 |
| chr16_-_25368048 | 0.32 |
ENSDART00000132445
|
rbfa
|
ribosome binding factor A |
| chr20_-_38746889 | 0.32 |
ENSDART00000140275
|
trim54
|
tripartite motif containing 54 |
| chr18_-_6534516 | 0.32 |
ENSDART00000009217
|
ddx11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr25_+_34407740 | 0.31 |
ENSDART00000012677
|
OTUD7A
|
si:dkey-37f18.2 |
| chr6_-_31576397 | 0.31 |
ENSDART00000111837
|
raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr5_-_68244564 | 0.31 |
ENSDART00000169350
|
CABZ01083944.1
|
|
| chr19_-_25772980 | 0.31 |
ENSDART00000052393
|
pard6gb
|
par-6 family cell polarity regulator gamma b |
| chr24_-_38644937 | 0.31 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr21_+_21791799 | 0.31 |
ENSDART00000151759
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr2_+_43204919 | 0.31 |
ENSDART00000160077
ENSDART00000018729 ENSDART00000129134 ENSDART00000056402 |
pard3ab
|
par-3 family cell polarity regulator alpha, b |
| chr12_+_34854562 | 0.31 |
ENSDART00000130366
|
si:dkey-21c1.4
|
si:dkey-21c1.4 |
| chr10_-_35257458 | 0.30 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr18_+_44750170 | 0.30 |
ENSDART00000132775
ENSDART00000187468 |
si:dkey-224e22.2
|
si:dkey-224e22.2 |
| chr7_-_24644893 | 0.30 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr5_+_27583445 | 0.29 |
ENSDART00000136488
|
zmat4a
|
zinc finger, matrin-type 4a |
| chr20_-_29499363 | 0.29 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr12_-_10381282 | 0.29 |
ENSDART00000152788
|
mki67
|
marker of proliferation Ki-67 |
| chr22_+_24603930 | 0.29 |
ENSDART00000180240
ENSDART00000164256 |
CU655842.1
|
|
| chr20_+_9474841 | 0.29 |
ENSDART00000053847
|
rad51b
|
RAD51 paralog B |
| chr21_+_40695345 | 0.29 |
ENSDART00000143594
|
ccdc82
|
coiled-coil domain containing 82 |
| chr13_-_42530656 | 0.29 |
ENSDART00000084327
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr2_-_43124747 | 0.29 |
ENSDART00000188883
|
lrrc6
|
leucine rich repeat containing 6 |
| chr6_-_55399214 | 0.28 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr2_-_20120904 | 0.28 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr6_+_37625787 | 0.28 |
ENSDART00000065122
|
tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr20_-_2713120 | 0.28 |
ENSDART00000138753
ENSDART00000104606 |
rars2
|
arginyl-tRNA synthetase 2, mitochondrial (putative) |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 0.8 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 0.5 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.2 | 0.6 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 0.5 | GO:0034214 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.2 | 0.5 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.7 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 1.0 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 1.0 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.1 | 0.5 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 0.7 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.7 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.3 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.1 | 0.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.3 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.7 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.8 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.3 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 1.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.2 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.1 | 0.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.9 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.3 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.2 | GO:0042671 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.5 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.2 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.2 | GO:0071675 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 0.2 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.2 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) response to dexamethasone(GO:0071548) |
| 0.1 | 0.2 | GO:0031446 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.2 | GO:0043102 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.3 | GO:0060343 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.0 | 0.1 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.6 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.3 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.7 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.3 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.5 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.6 | GO:0006684 | sphingomyelin metabolic process(GO:0006684) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0018202 | histone H3-K36 methylation(GO:0010452) peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.3 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.4 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 1.4 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.3 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.0 | 1.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.5 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.1 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0071543 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.6 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:0072088 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.0 | 0.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.3 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.2 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.6 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.2 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.4 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.2 | GO:0035476 | angioblast cell migration(GO:0035476) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.2 | 0.6 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.5 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.4 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.3 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.7 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 0.5 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.5 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.3 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.7 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 1.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.8 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 1.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.0 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |