Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
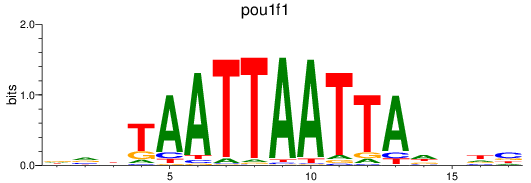
Results for pou1f1
Z-value: 0.82
Transcription factors associated with pou1f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou1f1
|
ENSDARG00000058924 | POU class 1 homeobox 1 |
|
pou1f1
|
ENSDARG00000110816 | POU class 1 homeobox 1 |
|
pou1f1
|
ENSDARG00000115109 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou1f1 | dr11_v1_chr9_-_19161982_19161982 | 0.87 | 2.5e-03 | Click! |
Activity profile of pou1f1 motif
Sorted Z-values of pou1f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22099536 | 3.72 |
ENSDART00000101923
|
CR391987.1
|
|
| chr4_+_9669717 | 3.48 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr12_-_17712393 | 3.46 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr3_+_28939759 | 2.65 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr9_-_22213297 | 2.32 |
ENSDART00000110656
ENSDART00000133149 |
crygm2d20
|
crystallin, gamma M2d20 |
| chr3_+_26145013 | 2.27 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr13_+_24279021 | 2.24 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr14_+_46313396 | 1.92 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr2_-_7666021 | 1.60 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr6_-_58764672 | 1.59 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr21_+_25236297 | 1.34 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr23_-_21453614 | 1.31 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr21_-_43015383 | 1.27 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr24_-_26328721 | 1.27 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr20_+_19512727 | 1.27 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr14_-_24761132 | 1.22 |
ENSDART00000146299
|
slit3
|
slit homolog 3 (Drosophila) |
| chr16_-_28856112 | 1.20 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr13_-_29420885 | 1.13 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr3_+_30922947 | 1.13 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr14_-_413273 | 1.10 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr24_+_32176155 | 1.08 |
ENSDART00000003745
|
vim
|
vimentin |
| chr5_-_71705191 | 1.07 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr21_+_45841731 | 1.04 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr5_+_71802014 | 1.03 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr9_-_42418470 | 1.03 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr5_-_41494831 | 1.02 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr14_-_4145594 | 0.96 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr3_+_36424055 | 0.96 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr8_+_7359294 | 0.96 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr4_-_2545310 | 0.95 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr7_+_59020972 | 0.92 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr15_+_9327252 | 0.92 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr6_-_11768198 | 0.92 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr7_+_19495379 | 0.91 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr12_+_27331324 | 0.90 |
ENSDART00000087208
|
sost
|
sclerostin |
| chr13_+_9432501 | 0.90 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr11_-_1509773 | 0.89 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr10_-_43771447 | 0.87 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr13_-_9886579 | 0.87 |
ENSDART00000101926
|
si:ch211-117n7.7
|
si:ch211-117n7.7 |
| chr16_-_21140097 | 0.86 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr9_+_34641237 | 0.85 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr19_+_12915498 | 0.85 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr23_+_19813677 | 0.84 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr2_+_2223837 | 0.83 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr19_+_43297546 | 0.82 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr17_-_30702411 | 0.81 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr23_-_21446985 | 0.81 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr20_-_5291012 | 0.79 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr5_-_44829719 | 0.79 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr23_-_42232124 | 0.79 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr7_+_71664624 | 0.78 |
ENSDART00000170273
|
emilin2b
|
elastin microfibril interfacer 2b |
| chr16_+_46111849 | 0.78 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr15_-_21014270 | 0.77 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr13_+_29770837 | 0.77 |
ENSDART00000076998
|
pax2a
|
paired box 2a |
| chr12_+_3022882 | 0.77 |
ENSDART00000122905
|
rac3b
|
Rac family small GTPase 3b |
| chr3_-_15999501 | 0.76 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr1_-_42289704 | 0.73 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr10_-_14556978 | 0.73 |
ENSDART00000126643
|
zgc:153395
|
zgc:153395 |
| chr24_+_21621654 | 0.73 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr10_+_18952271 | 0.72 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr23_-_23401305 | 0.71 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr6_+_52350443 | 0.71 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr14_-_24391424 | 0.70 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr7_+_26029672 | 0.69 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr23_+_31405497 | 0.69 |
ENSDART00000053546
|
sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr2_+_16160906 | 0.69 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr17_-_29224908 | 0.67 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr7_+_19495905 | 0.67 |
ENSDART00000125584
ENSDART00000173774 |
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr5_-_2282256 | 0.66 |
ENSDART00000064012
|
ca4a
|
carbonic anhydrase IV a |
| chr8_-_13574764 | 0.66 |
ENSDART00000076561
|
B3GNT3 (1 of many)
|
si:ch211-126g16.10 |
| chr12_-_558201 | 0.65 |
ENSDART00000168586
ENSDART00000158355 |
bsk146
|
brain specific kinase 146 |
| chr24_-_38657683 | 0.64 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr1_-_54063520 | 0.63 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr9_+_18716485 | 0.63 |
ENSDART00000135125
|
serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr12_+_13282797 | 0.62 |
ENSDART00000137757
ENSDART00000152397 |
irf9
|
interferon regulatory factor 9 |
| chr13_-_31452516 | 0.61 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr20_+_7584211 | 0.61 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr19_+_43359075 | 0.61 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr5_-_54672763 | 0.60 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr19_+_43780970 | 0.60 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr19_-_26869103 | 0.59 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr4_-_5302866 | 0.58 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr8_+_26141680 | 0.58 |
ENSDART00000078334
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr16_+_28728347 | 0.57 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr15_-_20939579 | 0.57 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr23_-_20051369 | 0.57 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr22_-_13851297 | 0.56 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr23_-_6522099 | 0.56 |
ENSDART00000092214
ENSDART00000183380 ENSDART00000138020 |
bmp7b
|
bone morphogenetic protein 7b |
| chr7_+_26534131 | 0.56 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr6_-_54815886 | 0.55 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr8_+_28259347 | 0.54 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr3_+_23092762 | 0.54 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr11_+_11974708 | 0.54 |
ENSDART00000125060
|
zgc:64002
|
zgc:64002 |
| chr11_+_30057762 | 0.53 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr11_+_40812590 | 0.53 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr3_-_19368435 | 0.53 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr14_-_17072736 | 0.53 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr16_+_28932038 | 0.53 |
ENSDART00000149480
|
npr1b
|
natriuretic peptide receptor 1b |
| chr4_+_22480169 | 0.52 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr17_-_12336987 | 0.52 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr20_+_4060839 | 0.52 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr13_-_31622195 | 0.51 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr22_-_10121880 | 0.50 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr16_-_12173554 | 0.50 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr1_+_51721851 | 0.50 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr1_+_5402476 | 0.50 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr21_-_20939488 | 0.50 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr19_+_10339538 | 0.50 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr3_-_36839115 | 0.49 |
ENSDART00000154553
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr8_+_24745041 | 0.48 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr1_+_25801648 | 0.48 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr25_+_31227747 | 0.48 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr11_+_43419809 | 0.48 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr16_-_28878080 | 0.47 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr9_+_7724152 | 0.47 |
ENSDART00000061716
|
mnx2a
|
motor neuron and pancreas homeobox 2a |
| chr15_-_46779934 | 0.47 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr8_-_2434282 | 0.47 |
ENSDART00000137262
ENSDART00000134044 |
vdac3
|
voltage-dependent anion channel 3 |
| chr23_+_45512825 | 0.46 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
| chr21_+_22840246 | 0.46 |
ENSDART00000151621
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr3_+_38540411 | 0.45 |
ENSDART00000154943
|
si:dkey-7f16.3
|
si:dkey-7f16.3 |
| chr24_+_9475809 | 0.44 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr13_-_2010191 | 0.43 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr22_+_30331414 | 0.43 |
ENSDART00000133482
|
BX649448.3
|
|
| chr21_-_25522510 | 0.43 |
ENSDART00000162711
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr4_+_25654686 | 0.43 |
ENSDART00000100714
|
acot16
|
acyl-CoA thioesterase 16 |
| chr1_+_44127292 | 0.42 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr18_+_24919614 | 0.42 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr16_+_11992498 | 0.41 |
ENSDART00000143442
|
p3h3
|
prolyl 3-hydroxylase 3 |
| chr21_+_19635486 | 0.41 |
ENSDART00000185736
|
fgf10a
|
fibroblast growth factor 10a |
| chr1_-_9980765 | 0.41 |
ENSDART00000142906
|
si:dkeyp-75b4.7
|
si:dkeyp-75b4.7 |
| chr16_+_12836143 | 0.41 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr1_-_31534089 | 0.41 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr10_-_33621739 | 0.41 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr22_-_31060579 | 0.41 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr23_-_18913032 | 0.41 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr6_+_6797520 | 0.40 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr6_-_50704689 | 0.40 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr2_+_20406399 | 0.40 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr8_+_8845932 | 0.39 |
ENSDART00000112028
|
si:ch211-180f4.1
|
si:ch211-180f4.1 |
| chr1_+_13087797 | 0.39 |
ENSDART00000170314
|
si:dkey-5n7.2
|
si:dkey-5n7.2 |
| chr3_-_19200571 | 0.39 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr16_+_36671064 | 0.38 |
ENSDART00000109703
|
col6a4a
|
collagen, type VI, alpha 4a |
| chr5_-_42083363 | 0.37 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr2_-_59327299 | 0.37 |
ENSDART00000133734
|
ftr36
|
finTRIM family, member 36 |
| chr20_+_25225112 | 0.37 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr1_-_22757145 | 0.37 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr1_+_31638274 | 0.36 |
ENSDART00000057885
|
fgf8b
|
fibroblast growth factor 8 b |
| chr11_+_30663300 | 0.36 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr10_-_35236949 | 0.35 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr12_+_1139690 | 0.35 |
ENSDART00000160442
|
CABZ01072885.1
|
|
| chr20_-_44557037 | 0.35 |
ENSDART00000140995
|
mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr4_+_5255041 | 0.34 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr5_-_65000312 | 0.34 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr19_-_32150078 | 0.34 |
ENSDART00000134934
ENSDART00000186410 ENSDART00000181780 |
pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr16_+_46401576 | 0.34 |
ENSDART00000130264
|
rpz
|
rapunzel |
| chr16_+_5408748 | 0.34 |
ENSDART00000160008
ENSDART00000014024 |
plecb
|
plectin b |
| chr12_+_39203745 | 0.34 |
ENSDART00000153661
|
si:dkeyp-106c3.2
|
si:dkeyp-106c3.2 |
| chr22_-_5529241 | 0.34 |
ENSDART00000157893
|
CABZ01064968.1
|
|
| chr8_+_52637507 | 0.34 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr17_-_32413147 | 0.34 |
ENSDART00000149102
|
grhl1
|
grainyhead-like transcription factor 1 |
| chr18_+_39937225 | 0.34 |
ENSDART00000141136
|
si:ch211-282k23.2
|
si:ch211-282k23.2 |
| chr16_+_9540033 | 0.33 |
ENSDART00000149574
|
ca14
|
carbonic anhydrase XIV |
| chr11_-_29737088 | 0.33 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr9_+_34127005 | 0.33 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr20_+_7180298 | 0.33 |
ENSDART00000143026
|
si:dkeyp-51f12.2
|
si:dkeyp-51f12.2 |
| chr7_+_65876335 | 0.33 |
ENSDART00000150143
|
tead1b
|
TEA domain family member 1b |
| chr2_-_59157790 | 0.32 |
ENSDART00000192303
ENSDART00000159362 |
ftr32
|
finTRIM family, member 32 |
| chr2_+_20496223 | 0.32 |
ENSDART00000157931
|
agla
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a |
| chr24_-_1151334 | 0.32 |
ENSDART00000039700
ENSDART00000177356 |
itgb1a
|
integrin, beta 1a |
| chr6_+_25257728 | 0.32 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr6_-_28980756 | 0.31 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr11_-_45141309 | 0.31 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr13_+_7442023 | 0.31 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr15_-_18162647 | 0.31 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr16_-_12173399 | 0.31 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr7_+_21859337 | 0.31 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr17_+_16564921 | 0.30 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr14_-_24081929 | 0.30 |
ENSDART00000158576
|
msx2a
|
muscle segment homeobox 2a |
| chr15_-_40157165 | 0.30 |
ENSDART00000192991
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr22_-_26865361 | 0.29 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr10_+_31248036 | 0.29 |
ENSDART00000193574
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr3_+_26342768 | 0.29 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr5_-_42904329 | 0.29 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr17_-_21418340 | 0.29 |
ENSDART00000007021
|
atp6v1ba
|
ATPase, H+ transporting, lysosomal, V1 subunit B, member a |
| chr6_+_40714811 | 0.29 |
ENSDART00000153868
|
ccdc36
|
coiled-coil domain containing 36 |
| chr6_+_58832323 | 0.29 |
ENSDART00000042595
|
dctn2
|
dynactin 2 (p50) |
| chr17_+_24851951 | 0.29 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr4_-_18309917 | 0.29 |
ENSDART00000189084
|
plxnc1
|
plexin C1 |
| chr1_-_22512063 | 0.29 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr20_+_11731039 | 0.28 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr18_-_47662696 | 0.28 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr5_+_65536095 | 0.28 |
ENSDART00000189898
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr3_+_23029934 | 0.28 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr24_-_7995960 | 0.28 |
ENSDART00000186594
|
bloc1s5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr9_+_55154414 | 0.28 |
ENSDART00000182924
|
ANOS1
|
anosmin 1 |
| chr10_+_2742499 | 0.28 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr14_+_4807207 | 0.28 |
ENSDART00000167145
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr15_-_40157331 | 0.28 |
ENSDART00000187958
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr12_+_31735159 | 0.28 |
ENSDART00000185442
|
RNF157
|
si:dkey-49c17.3 |
| chr13_+_24756486 | 0.27 |
ENSDART00000137074
|
col17a1b
|
collagen, type XVII, alpha 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou1f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0045988 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.4 | 1.3 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.3 | 1.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.3 | 0.8 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.2 | 0.9 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.2 | 1.6 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.2 | 0.9 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.2 | 1.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.5 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.5 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.4 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.5 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.5 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 2.6 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.9 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.4 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 1.8 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 0.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.8 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.5 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.4 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.5 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.8 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.3 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.1 | 0.6 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.3 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.4 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.7 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.2 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 1.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.5 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.5 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 1.3 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.0 | 1.0 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.5 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.8 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.7 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.8 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.8 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.9 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 4.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.3 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.3 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 1.2 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 2.2 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.6 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.3 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.8 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.3 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.9 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 1.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.8 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.0 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.8 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.4 | GO:0044259 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 0.2 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.3 | 1.0 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.3 | 2.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 0.8 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 0.9 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.2 | 2.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.4 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 2.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.8 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 0.4 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.9 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.9 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.8 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.3 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.1 | 0.4 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 1.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.3 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 1.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.3 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 4.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.0 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |