Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
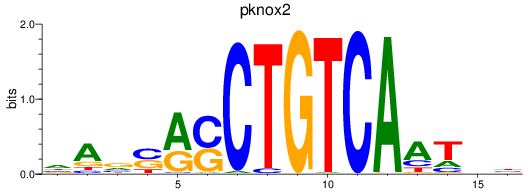
Results for pknox2
Z-value: 0.48
Transcription factors associated with pknox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pknox2
|
ENSDARG00000055349 | pbx/knotted 1 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pknox2 | dr11_v1_chr10_-_31175744_31175744 | 0.33 | 3.8e-01 | Click! |
Activity profile of pknox2 motif
Sorted Z-values of pknox2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_41468892 | 0.70 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr17_-_36860988 | 0.60 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr3_+_7771420 | 0.57 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr12_-_48168135 | 0.56 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr23_-_31060350 | 0.49 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr22_-_37834312 | 0.46 |
ENSDART00000076128
|
ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr8_-_32385989 | 0.46 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr2_-_11027258 | 0.45 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr14_+_30340251 | 0.44 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr19_-_25427255 | 0.44 |
ENSDART00000036854
|
glcci1a
|
glucocorticoid induced 1a |
| chr2_-_38117538 | 0.42 |
ENSDART00000013676
|
chd8
|
chromodomain helicase DNA binding protein 8 |
| chr23_-_36592436 | 0.40 |
ENSDART00000168246
|
spryd3
|
SPRY domain containing 3 |
| chr20_-_28433990 | 0.40 |
ENSDART00000182824
ENSDART00000193381 |
wdr21
|
WD repeat domain 21 |
| chr23_+_25893020 | 0.39 |
ENSDART00000144769
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr5_-_41124241 | 0.38 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr16_+_48714048 | 0.38 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr13_-_18011168 | 0.38 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr7_-_29723761 | 0.37 |
ENSDART00000173560
|
vps13c
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr11_-_15874974 | 0.37 |
ENSDART00000166551
ENSDART00000129526 ENSDART00000165836 |
rap1ab
|
RAP1A, member of RAS oncogene family b |
| chr8_+_44623540 | 0.37 |
ENSDART00000141513
|
grk5l
|
G protein-coupled receptor kinase 5 like |
| chr17_-_7792376 | 0.37 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr8_+_25959940 | 0.36 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr19_+_43905671 | 0.35 |
ENSDART00000168725
ENSDART00000133628 |
ankib1a
|
ankyrin repeat and IBR domain containing 1a |
| chr8_-_18613948 | 0.35 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr4_+_9279515 | 0.34 |
ENSDART00000048707
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr7_+_57089354 | 0.34 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr11_+_28218141 | 0.34 |
ENSDART00000043756
|
ephb2b
|
eph receptor B2b |
| chr4_+_9279784 | 0.34 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr7_-_37555208 | 0.33 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr5_+_27137473 | 0.32 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr20_+_21391181 | 0.32 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr3_+_14463941 | 0.32 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr13_-_7233811 | 0.32 |
ENSDART00000162026
|
ninl
|
ninein-like |
| chr17_+_27158706 | 0.31 |
ENSDART00000151829
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr23_+_13528053 | 0.30 |
ENSDART00000162217
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr20_+_27713210 | 0.30 |
ENSDART00000132222
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr16_-_42770064 | 0.30 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr22_+_25086942 | 0.29 |
ENSDART00000061117
|
rrbp1b
|
ribosome binding protein 1b |
| chr23_-_21758253 | 0.29 |
ENSDART00000046613
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr4_-_20081621 | 0.29 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr15_-_28587147 | 0.29 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr20_+_27712714 | 0.28 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr13_+_45524475 | 0.28 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr5_-_35200590 | 0.27 |
ENSDART00000051271
|
fcho2
|
FCH domain only 2 |
| chr4_+_26056548 | 0.27 |
ENSDART00000171204
|
SCYL2
|
si:ch211-244b2.1 |
| chr25_+_14870043 | 0.27 |
ENSDART00000035714
ENSDART00000171835 |
dnajc24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr20_-_20270191 | 0.26 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr22_+_25086567 | 0.26 |
ENSDART00000192114
ENSDART00000177284 ENSDART00000180296 ENSDART00000190384 |
rrbp1b
|
ribosome binding protein 1b |
| chr11_+_44503774 | 0.26 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr11_+_5880562 | 0.26 |
ENSDART00000129663
ENSDART00000130768 ENSDART00000160909 |
dazap1
|
DAZ associated protein 1 |
| chr24_+_33392698 | 0.25 |
ENSDART00000122579
|
si:ch73-173p19.1
|
si:ch73-173p19.1 |
| chr7_+_30725759 | 0.25 |
ENSDART00000127131
|
mtmr10
|
myotubularin related protein 10 |
| chr20_-_28433616 | 0.25 |
ENSDART00000169289
|
wdr21
|
WD repeat domain 21 |
| chr7_-_19369002 | 0.24 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr13_+_22712406 | 0.23 |
ENSDART00000132847
|
si:ch211-134m17.9
|
si:ch211-134m17.9 |
| chr10_+_37500234 | 0.23 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr9_+_27876146 | 0.23 |
ENSDART00000133997
|
armc8
|
armadillo repeat containing 8 |
| chr19_-_6134802 | 0.23 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr23_+_39963599 | 0.23 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr2_-_45510699 | 0.22 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr17_+_33999630 | 0.22 |
ENSDART00000167085
ENSDART00000155030 ENSDART00000168522 ENSDART00000191799 ENSDART00000189684 ENSDART00000153942 ENSDART00000187272 ENSDART00000127692 |
gphna
|
gephyrin a |
| chr10_+_28160265 | 0.22 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr21_+_39336285 | 0.21 |
ENSDART00000139677
|
si:ch211-274p24.4
|
si:ch211-274p24.4 |
| chr25_-_8138122 | 0.21 |
ENSDART00000104659
|
sergef
|
secretion regulating guanine nucleotide exchange factor |
| chr21_-_588858 | 0.20 |
ENSDART00000168983
|
TMEM38B
|
transmembrane protein 38B |
| chr5_-_23675222 | 0.20 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr6_+_59854224 | 0.20 |
ENSDART00000083499
|
kdm6al
|
lysine (K)-specific demethylase 6A, like |
| chr10_+_20590190 | 0.20 |
ENSDART00000131819
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr17_-_14780578 | 0.20 |
ENSDART00000154690
|
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr15_+_23528310 | 0.20 |
ENSDART00000152523
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr25_-_21894317 | 0.20 |
ENSDART00000089642
|
fbxo31
|
F-box protein 31 |
| chr5_-_18911114 | 0.20 |
ENSDART00000014434
|
bri3bp
|
bri3 binding protein |
| chr4_+_25693463 | 0.20 |
ENSDART00000132864
|
acot18
|
acyl-CoA thioesterase 18 |
| chr7_+_52761841 | 0.20 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr15_+_23528010 | 0.20 |
ENSDART00000152786
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr21_-_34658266 | 0.19 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr5_+_6796291 | 0.19 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr12_-_33659328 | 0.19 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr3_-_25646149 | 0.19 |
ENSDART00000122735
|
usp43b
|
ubiquitin specific peptidase 43b |
| chr14_-_21238046 | 0.19 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr19_+_42847306 | 0.19 |
ENSDART00000135164
|
pdcd6ip
|
programmed cell death 6 interacting protein |
| chr7_-_56766100 | 0.19 |
ENSDART00000189934
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr10_+_20589969 | 0.19 |
ENSDART00000183042
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr5_+_30384554 | 0.18 |
ENSDART00000135483
|
zgc:158412
|
zgc:158412 |
| chr16_+_11151699 | 0.18 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr3_-_23596809 | 0.17 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr11_-_843811 | 0.17 |
ENSDART00000173331
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr5_-_13766651 | 0.16 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr2_+_30489846 | 0.16 |
ENSDART00000145732
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr5_-_32489796 | 0.16 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr12_-_22039350 | 0.16 |
ENSDART00000153187
|
thrab
|
thyroid hormone receptor alpha b |
| chr18_+_22793743 | 0.15 |
ENSDART00000150106
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr16_-_31622777 | 0.15 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr3_-_20793655 | 0.15 |
ENSDART00000163473
ENSDART00000159457 |
spop
|
speckle-type POZ protein |
| chr19_-_30800004 | 0.14 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr14_+_29941266 | 0.13 |
ENSDART00000112757
|
fam149a
|
family with sequence similarity 149 member A |
| chr14_+_49152341 | 0.13 |
ENSDART00000084114
|
nsd1a
|
nuclear receptor binding SET domain protein 1a |
| chr22_-_17677947 | 0.13 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr1_-_19431510 | 0.13 |
ENSDART00000089025
|
lap3
|
leucine aminopeptidase 3 |
| chr23_+_1661743 | 0.13 |
ENSDART00000044776
|
stxbp3
|
syntaxin binding protein 3 |
| chr22_-_22231720 | 0.13 |
ENSDART00000160165
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr7_+_30725473 | 0.13 |
ENSDART00000085716
|
mtmr10
|
myotubularin related protein 10 |
| chr23_-_19103855 | 0.13 |
ENSDART00000016901
|
pard6b
|
par-6 partitioning defective 6 homolog beta (C. elegans) |
| chr3_+_13190012 | 0.13 |
ENSDART00000179747
ENSDART00000109876 ENSDART00000124824 ENSDART00000130261 |
sun1
|
Sad1 and UNC84 domain containing 1 |
| chr19_+_42693855 | 0.13 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr25_+_19008497 | 0.12 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr9_+_33267211 | 0.12 |
ENSDART00000025635
|
usp9
|
ubiquitin specific peptidase 9 |
| chr17_-_11418513 | 0.12 |
ENSDART00000064412
ENSDART00000151847 |
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr1_+_19849168 | 0.12 |
ENSDART00000111454
|
si:dkey-82j4.2
|
si:dkey-82j4.2 |
| chr8_+_45294767 | 0.11 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr22_+_18316144 | 0.11 |
ENSDART00000137985
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr2_+_394166 | 0.10 |
ENSDART00000155733
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr15_-_20528494 | 0.10 |
ENSDART00000048423
|
timm50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr11_-_19598809 | 0.10 |
ENSDART00000110499
|
atxn7
|
ataxin 7 |
| chr24_+_29352039 | 0.10 |
ENSDART00000101641
|
prmt6
|
protein arginine methyltransferase 6 |
| chr7_-_56766973 | 0.10 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr20_+_329032 | 0.09 |
ENSDART00000036635
|
fynb
|
FYN proto-oncogene, Src family tyrosine kinase b |
| chr24_-_26369185 | 0.09 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr3_-_40836081 | 0.09 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr13_+_33655404 | 0.08 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr24_-_21511737 | 0.08 |
ENSDART00000109587
|
rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr16_-_52646789 | 0.08 |
ENSDART00000035761
|
ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr12_+_3871452 | 0.08 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr19_+_25465025 | 0.07 |
ENSDART00000018553
|
rpa3
|
replication protein A3 |
| chr17_-_11417904 | 0.07 |
ENSDART00000103228
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr2_-_41723165 | 0.07 |
ENSDART00000155577
|
zgc:110158
|
zgc:110158 |
| chr4_+_16787040 | 0.06 |
ENSDART00000039027
|
golt1ba
|
golgi transport 1Ba |
| chr9_-_12885201 | 0.06 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr8_+_21437908 | 0.06 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr2_+_15128418 | 0.06 |
ENSDART00000141921
|
arhgap29b
|
Rho GTPase activating protein 29b |
| chr5_+_72145468 | 0.06 |
ENSDART00000148626
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr2_-_41723487 | 0.06 |
ENSDART00000170171
|
zgc:110158
|
zgc:110158 |
| chr3_-_15889508 | 0.06 |
ENSDART00000148363
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr7_-_28565230 | 0.06 |
ENSDART00000028887
|
tmem9b
|
TMEM9 domain family, member B |
| chr3_-_23596532 | 0.05 |
ENSDART00000124921
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr5_+_57743815 | 0.05 |
ENSDART00000005090
|
alg9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr14_-_689841 | 0.05 |
ENSDART00000125969
|
ube2ka
|
ubiquitin-conjugating enzyme E2Ka (UBC1 homolog, yeast) |
| chr6_-_23931442 | 0.05 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr5_-_13645995 | 0.04 |
ENSDART00000099665
ENSDART00000166957 |
purba
|
purine-rich element binding protein Ba |
| chr12_+_18899396 | 0.04 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr5_+_22307605 | 0.04 |
ENSDART00000138154
|
arhgap20b
|
Rho GTPase activating protein 20b |
| chr13_+_35765317 | 0.04 |
ENSDART00000100156
ENSDART00000167650 |
agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr21_+_43882274 | 0.04 |
ENSDART00000075672
|
sra1
|
steroid receptor RNA activator 1 |
| chr25_+_3549401 | 0.04 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr11_-_10456387 | 0.04 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr23_-_29667544 | 0.04 |
ENSDART00000059339
|
clstn1
|
calsyntenin 1 |
| chr9_+_34151367 | 0.03 |
ENSDART00000143991
|
gpr161
|
G protein-coupled receptor 161 |
| chr8_-_33114202 | 0.03 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr24_-_12981423 | 0.03 |
ENSDART00000133166
ENSDART00000131370 ENSDART00000002992 |
dcaf11
|
ddb1 and cul4 associated factor 11 |
| chr21_+_26539157 | 0.03 |
ENSDART00000021121
|
stx5al
|
syntaxin 5A, like |
| chr5_+_19320554 | 0.02 |
ENSDART00000165119
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr9_+_6009077 | 0.02 |
ENSDART00000057484
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr19_-_81477 | 0.02 |
ENSDART00000159815
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr6_+_41452979 | 0.02 |
ENSDART00000007353
|
wdr82
|
WD repeat domain 82 |
| chr7_-_26262978 | 0.01 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr25_+_32525131 | 0.01 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr13_-_42749916 | 0.01 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr10_+_36695597 | 0.01 |
ENSDART00000169015
ENSDART00000171392 |
rab6a
|
RAB6A, member RAS oncogene family |
| chr20_-_7547080 | 0.01 |
ENSDART00000146135
|
usp24
|
ubiquitin specific peptidase 24 |
| chr12_-_29301022 | 0.01 |
ENSDART00000187826
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr11_+_44356504 | 0.00 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr18_+_17786548 | 0.00 |
ENSDART00000189493
ENSDART00000146133 |
ZNF423
|
si:ch211-216l23.1 |
| chr18_+_17786710 | 0.00 |
ENSDART00000190203
ENSDART00000187095 ENSDART00000083296 |
ZNF423
|
si:ch211-216l23.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pknox2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.4 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.2 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.4 | GO:0071320 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.4 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.4 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.4 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.2 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |