Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
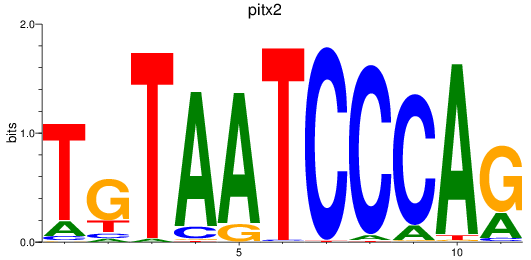
Results for pitx2
Z-value: 1.55
Transcription factors associated with pitx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pitx2
|
ENSDARG00000036194 | paired-like homeodomain 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pitx2 | dr11_v1_chr14_+_36218072_36218072 | 0.87 | 2.5e-03 | Click! |
Activity profile of pitx2 motif
Sorted Z-values of pitx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_45282858 | 7.64 |
ENSDART00000162353
|
CABZ01073265.1
|
|
| chr19_-_5345930 | 3.46 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr13_+_24280380 | 2.94 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr2_-_31767827 | 2.65 |
ENSDART00000114928
|
and2
|
actinodin2 |
| chr1_+_7546259 | 2.50 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr16_-_45910050 | 2.35 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr25_+_21833287 | 2.29 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr4_+_6643421 | 2.28 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr22_-_23625351 | 2.26 |
ENSDART00000192588
ENSDART00000163423 |
cfhl4
|
complement factor H like 4 |
| chr7_+_20505311 | 2.22 |
ENSDART00000187335
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr12_-_35949936 | 2.14 |
ENSDART00000192583
|
AL954682.1
|
|
| chr5_+_37978501 | 2.06 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr6_-_54126463 | 2.04 |
ENSDART00000161059
|
tusc2a
|
tumor suppressor candidate 2a |
| chr21_+_20715020 | 1.99 |
ENSDART00000015224
|
gadd45gb.1
|
growth arrest and DNA-damage-inducible, gamma b, tandem duplicate 1 |
| chr7_-_8408014 | 1.87 |
ENSDART00000112492
|
zgc:194686
|
zgc:194686 |
| chr11_+_6116503 | 1.86 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr1_+_31942961 | 1.79 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr6_+_29217392 | 1.78 |
ENSDART00000006386
|
atp1b1a
|
ATPase Na+/K+ transporting subunit beta 1a |
| chr23_-_45682136 | 1.77 |
ENSDART00000164646
|
fam160a1b
|
family with sequence similarity 160, member A1b |
| chr5_-_20205075 | 1.68 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr14_+_6423973 | 1.66 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr19_+_10339538 | 1.66 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr18_-_46354269 | 1.62 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr9_+_307863 | 1.58 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr20_-_29482492 | 1.58 |
ENSDART00000178308
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr24_+_36636208 | 1.58 |
ENSDART00000139211
|
si:ch73-334d15.4
|
si:ch73-334d15.4 |
| chr19_-_15855427 | 1.57 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr16_-_28836523 | 1.53 |
ENSDART00000108596
ENSDART00000142920 |
plin6
|
perilipin 6 |
| chr8_-_22964486 | 1.50 |
ENSDART00000112381
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr1_+_33322555 | 1.50 |
ENSDART00000113486
|
mxra5a
|
matrix-remodelling associated 5a |
| chr12_+_316238 | 1.50 |
ENSDART00000187492
|
rcvrnb
|
recoverin b |
| chr2_-_2020044 | 1.49 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr6_+_2093206 | 1.42 |
ENSDART00000114314
|
tgm2b
|
transglutaminase 2b |
| chr7_+_24115082 | 1.40 |
ENSDART00000182718
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr5_-_72125551 | 1.38 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr23_+_28731379 | 1.37 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr15_-_20024205 | 1.34 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr25_+_21832938 | 1.30 |
ENSDART00000148299
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr5_-_46505691 | 1.30 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr13_-_30028103 | 1.29 |
ENSDART00000183889
|
scdb
|
stearoyl-CoA desaturase b |
| chr25_+_6306885 | 1.28 |
ENSDART00000142705
ENSDART00000067510 |
crabp1a
|
cellular retinoic acid binding protein 1a |
| chr17_+_996509 | 1.25 |
ENSDART00000158830
|
cyp1c2
|
cytochrome P450, family 1, subfamily C, polypeptide 2 |
| chr19_+_19747430 | 1.25 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr14_-_2270973 | 1.24 |
ENSDART00000180729
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr24_+_24461341 | 1.22 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr12_+_9542124 | 1.21 |
ENSDART00000127952
|
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr19_-_28283844 | 1.21 |
ENSDART00000151756
ENSDART00000079104 |
ndufs6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr6_-_9581949 | 1.21 |
ENSDART00000144335
|
cyp27c1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr17_-_5583345 | 1.20 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr8_-_54304381 | 1.16 |
ENSDART00000184177
|
RHO (1 of many)
|
rhodopsin |
| chr7_-_57933736 | 1.15 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr11_+_5926850 | 1.15 |
ENSDART00000104364
|
rps15
|
ribosomal protein S15 |
| chr25_-_28384954 | 1.15 |
ENSDART00000073500
|
ptprz1a
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1a |
| chr15_-_21014015 | 1.15 |
ENSDART00000144991
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr15_+_35043007 | 1.13 |
ENSDART00000086954
|
sesn3
|
sestrin 3 |
| chr13_-_16222388 | 1.12 |
ENSDART00000182861
|
zgc:110045
|
zgc:110045 |
| chr10_-_43771447 | 1.12 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr17_-_31659670 | 1.12 |
ENSDART00000030448
|
vsx2
|
visual system homeobox 2 |
| chr10_+_10351685 | 1.09 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr10_+_21511495 | 1.07 |
ENSDART00000178395
|
BX957322.3
|
|
| chr7_+_24114694 | 1.05 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr21_+_41743493 | 1.03 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr24_+_20373605 | 1.02 |
ENSDART00000112349
|
plcd1a
|
phospholipase C, delta 1a |
| chr25_+_10458990 | 0.99 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr15_-_25584888 | 0.98 |
ENSDART00000127571
|
si:dkey-54n8.2
|
si:dkey-54n8.2 |
| chr11_+_6116096 | 0.97 |
ENSDART00000159680
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr23_-_15330168 | 0.96 |
ENSDART00000035865
ENSDART00000143635 |
sulf2b
|
sulfatase 2b |
| chr7_+_15872357 | 0.96 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr20_-_20533865 | 0.95 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr16_-_46660680 | 0.95 |
ENSDART00000159209
ENSDART00000191929 |
tmem176l.4
|
transmembrane protein 176l.4 |
| chr23_+_3607305 | 0.94 |
ENSDART00000186190
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr7_-_29021757 | 0.94 |
ENSDART00000086905
|
nrn1lb
|
neuritin 1-like b |
| chr23_+_3591690 | 0.94 |
ENSDART00000180822
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr18_+_6054816 | 0.94 |
ENSDART00000113668
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr8_-_22965214 | 0.92 |
ENSDART00000148178
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr18_+_507618 | 0.92 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr9_+_34127005 | 0.92 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr23_+_3627379 | 0.92 |
ENSDART00000187279
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr6_-_58149165 | 0.91 |
ENSDART00000170752
|
tox2
|
TOX high mobility group box family member 2 |
| chr8_-_18239494 | 0.91 |
ENSDART00000079989
ENSDART00000022959 |
guk1b
|
guanylate kinase 1b |
| chr2_-_17947389 | 0.91 |
ENSDART00000190089
ENSDART00000191872 ENSDART00000184039 ENSDART00000179791 |
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr23_+_3609535 | 0.90 |
ENSDART00000191440
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr15_-_21014270 | 0.90 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr1_-_52494122 | 0.90 |
ENSDART00000131407
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr20_-_49657134 | 0.89 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr13_-_24448278 | 0.89 |
ENSDART00000057584
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr23_+_3618454 | 0.88 |
ENSDART00000189393
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr12_+_48216662 | 0.87 |
ENSDART00000187369
|
lrrc20
|
leucine rich repeat containing 20 |
| chr23_+_43255328 | 0.87 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr18_-_40509006 | 0.87 |
ENSDART00000021372
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr9_-_23747264 | 0.85 |
ENSDART00000141461
ENSDART00000010311 |
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr10_-_43294933 | 0.85 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr20_+_36233873 | 0.85 |
ENSDART00000131867
|
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr22_+_5687615 | 0.85 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr10_+_15777064 | 0.84 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr6_+_21740672 | 0.83 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr3_-_25119839 | 0.82 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr19_-_9829965 | 0.82 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr15_-_12319065 | 0.82 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr19_-_3193912 | 0.80 |
ENSDART00000133159
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr3_+_37574885 | 0.80 |
ENSDART00000055225
|
wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr10_+_15777258 | 0.79 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr19_+_12762887 | 0.79 |
ENSDART00000139909
|
mc5ra
|
melanocortin 5a receptor |
| chr8_+_17184602 | 0.78 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr16_+_28383758 | 0.78 |
ENSDART00000059038
ENSDART00000141061 |
itga8
|
integrin, alpha 8 |
| chr18_+_507835 | 0.78 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr9_-_42989297 | 0.78 |
ENSDART00000126871
|
ttn.2
|
titin, tandem duplicate 2 |
| chr16_+_42018367 | 0.78 |
ENSDART00000058613
|
fli1b
|
Fli-1 proto-oncogene, ETS transcription factor b |
| chr21_+_19648814 | 0.78 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr23_+_9220436 | 0.78 |
ENSDART00000033663
ENSDART00000139870 |
rps21
|
ribosomal protein S21 |
| chr23_+_3616224 | 0.76 |
ENSDART00000190917
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr5_+_52067723 | 0.76 |
ENSDART00000166902
|
setbp1
|
SET binding protein 1 |
| chr25_-_13839743 | 0.75 |
ENSDART00000158780
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr6_+_37894914 | 0.75 |
ENSDART00000148817
|
oca2
|
oculocutaneous albinism II |
| chr23_+_3611765 | 0.75 |
ENSDART00000181481
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr7_+_57866292 | 0.75 |
ENSDART00000138757
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr6_-_54179860 | 0.74 |
ENSDART00000164283
|
rps10
|
ribosomal protein S10 |
| chr9_+_21401189 | 0.73 |
ENSDART00000062669
|
cx30.3
|
connexin 30.3 |
| chr12_+_47917971 | 0.73 |
ENSDART00000185933
|
tbata
|
thymus, brain and testes associated |
| chr18_-_17001056 | 0.73 |
ENSDART00000112627
|
tph2
|
tryptophan hydroxylase 2 (tryptophan 5-monooxygenase) |
| chr23_+_37090889 | 0.72 |
ENSDART00000074406
|
ubxn10
|
UBX domain protein 10 |
| chr3_+_7001608 | 0.72 |
ENSDART00000127809
|
zmp:0000001228
|
zmp:0000001228 |
| chr23_+_3613994 | 0.71 |
ENSDART00000183763
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr23_+_3622920 | 0.70 |
ENSDART00000183091
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr19_-_18626515 | 0.70 |
ENSDART00000160624
|
rps18
|
ribosomal protein S18 |
| chr23_+_3620618 | 0.69 |
ENSDART00000190126
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr18_-_17000842 | 0.69 |
ENSDART00000079832
|
tph2
|
tryptophan hydroxylase 2 (tryptophan 5-monooxygenase) |
| chr19_+_37701450 | 0.68 |
ENSDART00000087694
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr9_-_32753535 | 0.68 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr11_-_44409856 | 0.68 |
ENSDART00000162886
|
il1rapl1b
|
interleukin 1 receptor accessory protein-like 1b |
| chr23_-_28239750 | 0.68 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr15_+_618081 | 0.68 |
ENSDART00000181518
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr13_-_30027730 | 0.66 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr5_+_17727310 | 0.66 |
ENSDART00000147657
|
fbrsl1
|
fibrosin-like 1 |
| chr11_-_36051004 | 0.66 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr6_+_39506043 | 0.65 |
ENSDART00000086260
|
CR388364.1
|
|
| chr8_+_45003659 | 0.65 |
ENSDART00000132663
|
si:ch211-163b2.4
|
si:ch211-163b2.4 |
| chr16_+_46695777 | 0.65 |
ENSDART00000169767
|
rab25b
|
RAB25, member RAS oncogene family b |
| chr9_-_9998087 | 0.64 |
ENSDART00000124423
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr19_-_31802296 | 0.64 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr12_-_30345444 | 0.64 |
ENSDART00000152985
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr23_+_3587230 | 0.63 |
ENSDART00000055103
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr8_+_27807974 | 0.63 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr23_+_3596401 | 0.63 |
ENSDART00000185908
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr13_-_40411908 | 0.62 |
ENSDART00000057094
ENSDART00000150091 |
nkx2.3
|
NK2 homeobox 3 |
| chr23_-_13295923 | 0.62 |
ENSDART00000189214
ENSDART00000165432 |
CR450832.1
|
Danio rerio uncharacterized LOC100001444 (LOC100001444), mRNA. |
| chr23_+_3602779 | 0.61 |
ENSDART00000013629
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr8_+_694218 | 0.61 |
ENSDART00000147753
|
rnf165b
|
ring finger protein 165b |
| chr1_-_26293203 | 0.61 |
ENSDART00000180140
|
cxxc4
|
CXXC finger 4 |
| chr11_-_4235811 | 0.60 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr9_-_9980704 | 0.60 |
ENSDART00000130243
ENSDART00000193475 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr13_+_50151407 | 0.59 |
ENSDART00000031858
|
gpr137ba
|
G protein-coupled receptor 137Ba |
| chr3_-_28828569 | 0.57 |
ENSDART00000167679
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr14_-_32884138 | 0.57 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr7_-_22132265 | 0.57 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr23_+_3598320 | 0.57 |
ENSDART00000189608
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr14_+_9009600 | 0.56 |
ENSDART00000133904
|
si:ch211-274f20.2
|
si:ch211-274f20.2 |
| chr3_+_16762483 | 0.56 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr2_+_36701322 | 0.56 |
ENSDART00000002510
|
golim4b
|
golgi integral membrane protein 4b |
| chr9_-_1986014 | 0.54 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr21_+_10756154 | 0.54 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr25_+_35019693 | 0.54 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr23_+_3594171 | 0.53 |
ENSDART00000159609
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr12_+_30653047 | 0.53 |
ENSDART00000148562
|
thbs2b
|
thrombospondin 2b |
| chr15_-_23467750 | 0.52 |
ENSDART00000148804
|
pdzd3a
|
PDZ domain containing 3a |
| chr3_+_12334509 | 0.52 |
ENSDART00000165796
|
glis2b
|
GLIS family zinc finger 2b |
| chr5_-_38094130 | 0.52 |
ENSDART00000131831
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr24_-_27400630 | 0.52 |
ENSDART00000165760
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr12_+_17106117 | 0.51 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr21_+_22630627 | 0.51 |
ENSDART00000193092
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr23_+_3589711 | 0.51 |
ENSDART00000187435
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr23_+_3600866 | 0.51 |
ENSDART00000184376
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr18_+_33725576 | 0.51 |
ENSDART00000146816
|
si:dkey-145c18.5
|
si:dkey-145c18.5 |
| chr23_+_3625083 | 0.51 |
ENSDART00000184958
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr5_+_64840656 | 0.50 |
ENSDART00000073953
|
lrrc8ab
|
leucine rich repeat containing 8 VRAC subunit Ab |
| chr20_-_25369767 | 0.50 |
ENSDART00000180278
|
itsn2a
|
intersectin 2a |
| chr22_-_18491813 | 0.49 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr23_+_3605325 | 0.49 |
ENSDART00000182713
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr25_+_26844028 | 0.49 |
ENSDART00000127274
ENSDART00000156179 |
sema7a
|
semaphorin 7A |
| chr11_+_45422206 | 0.49 |
ENSDART00000182548
|
hrasls
|
HRAS-like suppressor |
| chr6_-_39344259 | 0.48 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr4_-_9764767 | 0.48 |
ENSDART00000164328
ENSDART00000147699 |
mical3b
|
microtubule associated monooxygenase, calponin and LIM domain containing 3b |
| chr1_-_26292897 | 0.48 |
ENSDART00000112899
ENSDART00000185410 |
cxxc4
|
CXXC finger 4 |
| chr22_+_696931 | 0.47 |
ENSDART00000149712
ENSDART00000009756 |
gpr37l1a
|
G protein-coupled receptor 37 like 1a |
| chr7_+_30970045 | 0.47 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr21_+_22630297 | 0.46 |
ENSDART00000147175
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr9_-_43142636 | 0.46 |
ENSDART00000134349
ENSDART00000181835 |
ccdc141
|
coiled-coil domain containing 141 |
| chr23_+_9867483 | 0.45 |
ENSDART00000023099
|
slc16a7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2) |
| chr11_-_29563437 | 0.45 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr5_-_67145505 | 0.44 |
ENSDART00000011295
|
rom1a
|
retinal outer segment membrane protein 1a |
| chr16_+_26863414 | 0.43 |
ENSDART00000140673
|
galnt1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 |
| chr24_-_21258945 | 0.43 |
ENSDART00000111025
|
boc
|
BOC cell adhesion associated, oncogene regulated |
| chr23_+_3585251 | 0.43 |
ENSDART00000187356
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr23_+_13814978 | 0.43 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr6_-_36182115 | 0.42 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr19_-_40186328 | 0.42 |
ENSDART00000087474
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr19_-_25271155 | 0.41 |
ENSDART00000104027
|
rims3
|
regulating synaptic membrane exocytosis 3 |
| chr18_-_40508528 | 0.41 |
ENSDART00000185249
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr14_-_2264494 | 0.41 |
ENSDART00000191149
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr1_+_50613868 | 0.41 |
ENSDART00000111114
|
HERC5
|
si:ch73-190m4.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pitx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.4 | 1.7 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.4 | 2.1 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.4 | 2.0 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.3 | 1.7 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.3 | 1.6 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.3 | 1.5 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.3 | 3.6 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.3 | 1.6 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.3 | 1.3 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.3 | 1.8 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.2 | 1.0 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 2.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 1.4 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.2 | 1.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 0.8 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.2 | 0.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 0.8 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.2 | 0.5 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.2 | 0.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 0.7 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.2 | 1.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 0.5 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.2 | 1.1 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.2 | 1.7 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.6 | GO:0015865 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.8 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 1.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 2.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.8 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 14.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 1.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 2.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.9 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.3 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.1 | 1.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 1.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.7 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 0.8 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 2.4 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.8 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 2.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.3 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.8 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 1.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 1.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 2.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 1.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 0.3 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.1 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.8 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.5 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.7 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.2 | GO:0072012 | renal system vasculature development(GO:0061437) kidney vasculature development(GO:0061440) glomerulus vasculature development(GO:0072012) |
| 0.0 | 0.5 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.4 | GO:1902882 | regulation of response to oxidative stress(GO:1902882) |
| 0.0 | 0.9 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 2.9 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.2 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.3 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.5 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.7 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 2.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.9 | GO:0007599 | hemostasis(GO:0007599) |
| 0.0 | 1.3 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 2.0 | GO:0033333 | fin development(GO:0033333) |
| 0.0 | 0.5 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.8 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.8 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 1.3 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.5 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 1.0 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.2 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 5.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 1.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 2.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 2.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.8 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 2.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 2.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 3.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.7 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 1.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.4 | 1.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.4 | 2.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 1.7 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 2.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 1.4 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.3 | 3.6 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.3 | 1.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 1.0 | GO:0032896 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 0.7 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.2 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 0.5 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.2 | 1.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 0.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.6 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.6 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 2.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 1.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.8 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.8 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.5 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.6 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 1.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 5.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 3.8 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.4 | GO:0005523 | actin monomer binding(GO:0003785) tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 2.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |