Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
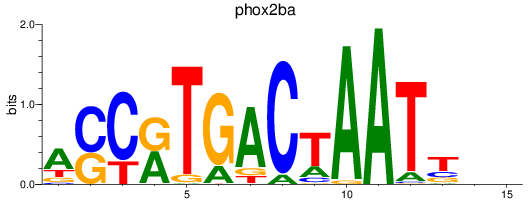
Results for phox2ba
Z-value: 0.95
Transcription factors associated with phox2ba
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
phox2ba
|
ENSDARG00000075330 | paired like homeobox 2Ba |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| phox2ba | dr11_v1_chr1_-_19079957_19079957 | -0.41 | 2.7e-01 | Click! |
Activity profile of phox2ba motif
Sorted Z-values of phox2ba motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_77551860 | 2.40 |
ENSDART00000188176
|
AL935186.6
|
|
| chr3_-_40054615 | 1.30 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr2_+_44512324 | 1.20 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr21_+_18910502 | 1.20 |
ENSDART00000147567
|
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr12_+_32368574 | 1.19 |
ENSDART00000086389
|
ANKFN1
|
si:ch211-277e21.2 |
| chr24_+_39227519 | 1.15 |
ENSDART00000184611
ENSDART00000193494 ENSDART00000190728 ENSDART00000168705 |
MPRIP
|
si:ch73-103b11.2 |
| chr9_+_22634073 | 1.09 |
ENSDART00000181822
|
etv5a
|
ets variant 5a |
| chr18_-_12612699 | 1.08 |
ENSDART00000090335
|
hipk2
|
homeodomain interacting protein kinase 2 |
| chr9_+_55321322 | 0.98 |
ENSDART00000111111
ENSDART00000191579 |
nlgn4b
|
neuroligin 4b |
| chr18_+_27598755 | 0.96 |
ENSDART00000193808
|
cd82b
|
CD82 molecule b |
| chr17_-_20202725 | 0.93 |
ENSDART00000133650
|
ecd
|
ecdysoneless homolog (Drosophila) |
| chr3_+_23247325 | 0.91 |
ENSDART00000114190
|
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr8_+_10862353 | 0.89 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr15_-_23908605 | 0.82 |
ENSDART00000185578
|
usp32
|
ubiquitin specific peptidase 32 |
| chr5_+_61941610 | 0.81 |
ENSDART00000168808
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr16_-_25519762 | 0.81 |
ENSDART00000146479
ENSDART00000142062 |
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr13_-_34862452 | 0.80 |
ENSDART00000134573
ENSDART00000047552 |
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr16_+_53387085 | 0.80 |
ENSDART00000154223
ENSDART00000101404 |
kif13a
|
kinesin family member 13A |
| chr15_+_25681044 | 0.78 |
ENSDART00000077853
|
hic1
|
hypermethylated in cancer 1 |
| chr1_+_26677406 | 0.75 |
ENSDART00000183427
ENSDART00000180366 ENSDART00000181997 |
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr5_-_25236340 | 0.73 |
ENSDART00000162774
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr7_-_41746427 | 0.73 |
ENSDART00000174232
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr8_-_11835607 | 0.71 |
ENSDART00000183601
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr13_-_12021566 | 0.68 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr12_-_30043809 | 0.65 |
ENSDART00000150220
ENSDART00000075889 ENSDART00000148847 |
trub1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr1_+_26676758 | 0.64 |
ENSDART00000152299
|
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr8_-_38355268 | 0.63 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr14_+_31493306 | 0.62 |
ENSDART00000138341
|
phf6
|
PHD finger protein 6 |
| chr2_-_9971609 | 0.61 |
ENSDART00000137924
ENSDART00000048655 ENSDART00000131613 |
zgc:55943
|
zgc:55943 |
| chr8_+_10835456 | 0.61 |
ENSDART00000151388
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr13_+_42858228 | 0.61 |
ENSDART00000179763
|
cdh23
|
cadherin-related 23 |
| chr8_-_11832771 | 0.59 |
ENSDART00000193412
ENSDART00000184720 ENSDART00000139947 |
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr2_-_26590628 | 0.57 |
ENSDART00000025120
|
ndc1
|
NDC1 transmembrane nucleoporin |
| chr17_+_27134806 | 0.56 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr11_+_24800156 | 0.56 |
ENSDART00000131976
|
adipor1a
|
adiponectin receptor 1a |
| chr16_+_1471 | 0.55 |
ENSDART00000181972
|
CABZ01090785.1
|
|
| chr14_+_45702521 | 0.54 |
ENSDART00000112470
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr8_+_7854130 | 0.51 |
ENSDART00000165575
|
cxxc1a
|
CXXC finger protein 1a |
| chr18_+_27001115 | 0.48 |
ENSDART00000133547
|
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr18_+_27000850 | 0.46 |
ENSDART00000188906
ENSDART00000086131 |
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr24_+_19542323 | 0.44 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr11_-_23459779 | 0.43 |
ENSDART00000183935
ENSDART00000184125 ENSDART00000193284 |
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr6_+_33537267 | 0.40 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr24_-_25574967 | 0.38 |
ENSDART00000189828
|
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr14_-_47314011 | 0.36 |
ENSDART00000178523
|
fstl5
|
follistatin-like 5 |
| chr19_+_42047427 | 0.35 |
ENSDART00000180225
ENSDART00000145356 |
si:ch211-13c6.2
|
si:ch211-13c6.2 |
| chr21_+_44300689 | 0.34 |
ENSDART00000186298
ENSDART00000142810 |
gabra3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr1_+_10074601 | 0.33 |
ENSDART00000168473
|
si:ch73-132f6.5
|
si:ch73-132f6.5 |
| chr21_+_27189490 | 0.31 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr22_+_2533514 | 0.31 |
ENSDART00000147967
|
si:ch73-92e7.4
|
si:ch73-92e7.4 |
| chr6_-_11800427 | 0.31 |
ENSDART00000126243
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr12_-_28956921 | 0.30 |
ENSDART00000142933
|
znf646
|
zinc finger protein 646 |
| chr14_-_45702721 | 0.23 |
ENSDART00000165727
|
si:dkey-226n24.1
|
si:dkey-226n24.1 |
| chr7_+_72460911 | 0.21 |
ENSDART00000160682
ENSDART00000168532 |
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr6_+_16493755 | 0.21 |
ENSDART00000126318
|
si:ch73-193c12.2
|
si:ch73-193c12.2 |
| chr19_+_10799929 | 0.20 |
ENSDART00000159152
|
lipea
|
lipase, hormone-sensitive a |
| chr25_+_32530976 | 0.19 |
ENSDART00000156190
ENSDART00000103324 |
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr13_+_47623916 | 0.19 |
ENSDART00000109266
|
mertka
|
c-mer proto-oncogene tyrosine kinase a |
| chr20_-_38801981 | 0.18 |
ENSDART00000125333
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr15_+_2500510 | 0.14 |
ENSDART00000180329
|
wdr53
|
WD repeat domain 53 |
| chr11_+_13223625 | 0.14 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr14_+_32839535 | 0.12 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr10_+_26645953 | 0.12 |
ENSDART00000131482
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr1_+_33390286 | 0.11 |
ENSDART00000139004
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr2_-_51223658 | 0.06 |
ENSDART00000171770
|
pigrl4.2
|
polymeric immunoglobulin receptor-like 4.2 |
| chr1_+_58205904 | 0.02 |
ENSDART00000147324
|
si:dkey-222h21.12
|
si:dkey-222h21.12 |
| chr10_+_26646104 | 0.01 |
ENSDART00000187685
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr20_-_18736281 | 0.01 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr8_+_16758304 | 0.01 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
Network of associatons between targets according to the STRING database.
First level regulatory network of phox2ba
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 0.8 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.2 | 0.7 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.2 | 1.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.7 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.6 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 1.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.1 | 1.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 1.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.8 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.8 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.9 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.2 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.1 | 0.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.4 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.6 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.4 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.8 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.7 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.8 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.9 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.9 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.3 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |