Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for pgr
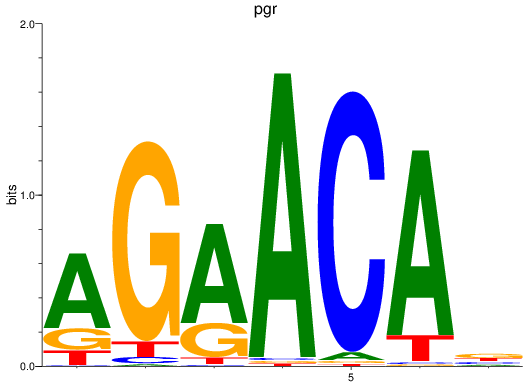
Z-value: 1.92
Transcription factors associated with pgr
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pgr
|
ENSDARG00000035966 | progesterone receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pgr | dr11_v1_chr18_+_41820039_41820039 | -0.32 | 4.0e-01 | Click! |
Activity profile of pgr motif
Sorted Z-values of pgr motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_21471022 | 3.60 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr23_+_21459263 | 3.20 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr9_-_22272181 | 3.17 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr25_+_31277415 | 3.15 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr23_-_21453614 | 2.72 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr18_+_9171778 | 2.55 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr23_-_45501177 | 2.50 |
ENSDART00000150103
|
col24a1
|
collagen type XXIV alpha 1 |
| chr5_-_32308526 | 2.42 |
ENSDART00000193758
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr9_-_22831836 | 2.39 |
ENSDART00000142585
|
neb
|
nebulin |
| chr16_+_23921777 | 2.35 |
ENSDART00000163213
|
apoa4b.3
|
apolipoprotein A-IV b, tandem duplicate 3 |
| chr14_-_25949713 | 2.30 |
ENSDART00000181455
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr2_-_29485408 | 2.24 |
ENSDART00000013411
|
cahz
|
carbonic anhydrase |
| chr4_-_16406737 | 2.19 |
ENSDART00000013085
|
dcn
|
decorin |
| chr23_+_21455152 | 2.16 |
ENSDART00000158511
ENSDART00000161321 ENSDART00000160731 ENSDART00000137573 |
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr9_-_33107237 | 2.14 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr5_-_20195350 | 2.14 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr21_+_25187210 | 2.09 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr20_+_16750177 | 1.93 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr5_-_67911111 | 1.92 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr3_-_28048475 | 1.88 |
ENSDART00000150888
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr19_-_42556086 | 1.85 |
ENSDART00000051731
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr6_-_40429411 | 1.80 |
ENSDART00000156005
ENSDART00000156357 |
si:dkey-28n18.9
|
si:dkey-28n18.9 |
| chr23_-_26077038 | 1.79 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr24_-_33756003 | 1.77 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr2_+_47581997 | 1.76 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr15_+_32710300 | 1.76 |
ENSDART00000167857
|
postnb
|
periostin, osteoblast specific factor b |
| chr10_-_29903165 | 1.76 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr10_+_21786656 | 1.73 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr18_+_16744307 | 1.71 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr7_-_74090168 | 1.71 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr8_+_25902170 | 1.68 |
ENSDART00000193130
|
rhoab
|
ras homolog gene family, member Ab |
| chr11_-_1024483 | 1.68 |
ENSDART00000017551
|
slc6a1b
|
solute carrier family 6 (neurotransmitter transporter), member 1b |
| chr22_-_26005894 | 1.68 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr1_+_44911405 | 1.66 |
ENSDART00000182465
|
wu:fc21g02
|
wu:fc21g02 |
| chr9_-_42696408 | 1.61 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr20_+_572037 | 1.59 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr22_-_23668356 | 1.58 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr18_-_6634424 | 1.58 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr17_-_6730247 | 1.58 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr22_-_26100282 | 1.58 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr20_+_2281933 | 1.57 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr15_-_23699737 | 1.56 |
ENSDART00000078311
|
zgc:154093
|
zgc:154093 |
| chr7_-_27033080 | 1.54 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr4_+_17417111 | 1.54 |
ENSDART00000056005
|
ascl1a
|
achaete-scute family bHLH transcription factor 1a |
| chr4_-_16333944 | 1.52 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr20_-_30377221 | 1.52 |
ENSDART00000126229
|
rps7
|
ribosomal protein S7 |
| chr24_-_27473771 | 1.48 |
ENSDART00000139874
|
cxl34b.11
|
CX chemokine ligand 34b, duplicate 11 |
| chr13_-_36391496 | 1.48 |
ENSDART00000100217
ENSDART00000140243 |
actn1
|
actinin, alpha 1 |
| chr24_+_23742690 | 1.46 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr14_-_36862745 | 1.46 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr6_-_39764995 | 1.46 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr5_-_51756210 | 1.45 |
ENSDART00000163464
|
lhfpl2b
|
LHFPL tetraspan subfamily member 2b |
| chr3_+_23731109 | 1.44 |
ENSDART00000131410
|
hoxb3a
|
homeobox B3a |
| chr14_-_2602445 | 1.43 |
ENSDART00000166910
|
etf1a
|
eukaryotic translation termination factor 1a |
| chr22_-_15602593 | 1.42 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr5_-_41307550 | 1.41 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr14_-_16328331 | 1.39 |
ENSDART00000111262
|
pcdh12
|
protocadherin 12 |
| chr17_+_26815021 | 1.36 |
ENSDART00000086885
|
asmt2
|
acetylserotonin O-methyltransferase 2 |
| chr15_+_37197494 | 1.33 |
ENSDART00000166203
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr8_+_23093155 | 1.33 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr22_-_15602760 | 1.32 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr4_+_18960914 | 1.31 |
ENSDART00000139730
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr24_+_3307857 | 1.31 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr1_-_5746030 | 1.30 |
ENSDART00000150863
|
nrp2a
|
neuropilin 2a |
| chr23_+_36095260 | 1.30 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr20_-_30376433 | 1.30 |
ENSDART00000190737
|
rps7
|
ribosomal protein S7 |
| chr6_+_23887314 | 1.30 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr7_+_39389273 | 1.29 |
ENSDART00000191298
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr15_+_32710094 | 1.28 |
ENSDART00000159592
|
postnb
|
periostin, osteoblast specific factor b |
| chr20_+_33904258 | 1.28 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr11_+_2202987 | 1.26 |
ENSDART00000190008
ENSDART00000173139 |
hoxc6b
|
homeobox C6b |
| chr9_-_46842179 | 1.26 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr3_+_7001608 | 1.25 |
ENSDART00000127809
|
zmp:0000001228
|
zmp:0000001228 |
| chr6_-_60031693 | 1.22 |
ENSDART00000160275
|
CABZ01079262.1
|
|
| chr1_-_20271138 | 1.20 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr10_+_26669177 | 1.20 |
ENSDART00000143402
|
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr6_+_41096058 | 1.19 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr9_-_18716 | 1.18 |
ENSDART00000164763
|
CABZ01078737.1
|
|
| chr22_-_24285432 | 1.16 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr24_-_38816725 | 1.14 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr2_+_29976419 | 1.13 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr13_-_31622195 | 1.12 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr21_-_25601648 | 1.12 |
ENSDART00000042578
|
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr20_+_29634653 | 1.12 |
ENSDART00000101556
|
asap2b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2b |
| chr14_-_25985698 | 1.12 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr10_+_31938796 | 1.11 |
ENSDART00000182579
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr19_-_2317558 | 1.11 |
ENSDART00000190300
|
sp8a
|
sp8 transcription factor a |
| chr7_+_25033924 | 1.11 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr16_-_36099492 | 1.11 |
ENSDART00000180905
|
CU499336.2
|
|
| chr5_-_28029558 | 1.11 |
ENSDART00000078649
|
abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr5_+_39667272 | 1.10 |
ENSDART00000085388
|
bmp3
|
bone morphogenetic protein 3 |
| chr3_-_58650057 | 1.10 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr19_+_19762183 | 1.10 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr20_-_29483514 | 1.09 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr2_+_47582681 | 1.09 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr20_+_539852 | 1.09 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr14_+_6159162 | 1.09 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr15_-_24869826 | 1.09 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr2_+_47582488 | 1.08 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr19_-_32641725 | 1.08 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr18_-_10995410 | 1.08 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr16_-_2414063 | 1.07 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr2_+_26288301 | 1.06 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr6_-_39765546 | 1.06 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr2_-_32352946 | 1.06 |
ENSDART00000144870
ENSDART00000077151 |
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr3_-_29977495 | 1.06 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr19_-_29437108 | 1.05 |
ENSDART00000052108
ENSDART00000074497 |
fndc5b
|
fibronectin type III domain containing 5b |
| chr4_-_5302162 | 1.05 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr11_+_21910343 | 1.04 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr19_+_48251273 | 1.04 |
ENSDART00000157424
|
CU693379.1
|
|
| chr14_-_2050057 | 1.04 |
ENSDART00000112875
|
PCDHB15
|
protocadherin beta 15 |
| chr3_+_20156956 | 1.04 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr3_-_26109322 | 1.03 |
ENSDART00000113780
|
zgc:162612
|
zgc:162612 |
| chr14_-_2189889 | 1.02 |
ENSDART00000181557
ENSDART00000106707 |
pcdh2ab9
pcdh2ab11
|
protocadherin 2 alpha b 9 protocadherin 2 alpha b 11 |
| chr15_-_35406564 | 1.01 |
ENSDART00000086051
|
mecom
|
MDS1 and EVI1 complex locus |
| chr15_+_28482862 | 1.01 |
ENSDART00000015286
ENSDART00000154320 |
ankrd13b
|
ankyrin repeat domain 13B |
| chr2_-_24996441 | 1.01 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr20_-_49657134 | 1.00 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr21_+_22985078 | 1.00 |
ENSDART00000156491
|
lpar6b
|
lysophosphatidic acid receptor 6b |
| chr21_-_26089964 | 1.00 |
ENSDART00000027848
|
tlcd1
|
TLC domain containing 1 |
| chr19_-_18626952 | 0.99 |
ENSDART00000168004
ENSDART00000162034 ENSDART00000165486 ENSDART00000167971 |
rps18
|
ribosomal protein S18 |
| chr20_+_6630540 | 0.99 |
ENSDART00000138361
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr1_-_35924495 | 0.98 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr16_-_35975254 | 0.98 |
ENSDART00000167537
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr23_+_21663631 | 0.98 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr9_+_3429662 | 0.97 |
ENSDART00000160977
ENSDART00000114168 ENSDART00000082153 |
CU469503.1
itga6a
|
integrin, alpha 6a |
| chr15_-_17870090 | 0.97 |
ENSDART00000155066
|
atf5b
|
activating transcription factor 5b |
| chr21_+_19648814 | 0.96 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr1_-_26398361 | 0.95 |
ENSDART00000160183
|
ppa2
|
pyrophosphatase (inorganic) 2 |
| chr12_-_1951233 | 0.95 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr5_+_20693724 | 0.95 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr4_+_77021784 | 0.95 |
ENSDART00000135345
ENSDART00000133855 |
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr4_-_14328997 | 0.95 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr15_+_1796313 | 0.93 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr7_+_73780936 | 0.93 |
ENSDART00000092691
|
pea15
|
proliferation and apoptosis adaptor protein 15 |
| chr19_-_47555956 | 0.93 |
ENSDART00000114549
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr21_+_28478663 | 0.92 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr1_+_19303241 | 0.91 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr2_-_37893910 | 0.91 |
ENSDART00000143027
|
hbl2
|
hexose-binding lectin 2 |
| chr22_-_968484 | 0.90 |
ENSDART00000105895
|
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr2_-_30611389 | 0.90 |
ENSDART00000142500
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr25_-_210730 | 0.89 |
ENSDART00000187580
|
FP236318.1
|
|
| chr4_-_11577253 | 0.89 |
ENSDART00000144452
|
net1
|
neuroepithelial cell transforming 1 |
| chr1_-_23274038 | 0.89 |
ENSDART00000181658
|
rfc1
|
replication factor C (activator 1) 1 |
| chr25_+_22274642 | 0.89 |
ENSDART00000127099
|
nr2e3
|
nuclear receptor subfamily 2, group E, member 3 |
| chr5_-_55395964 | 0.88 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr21_+_25643880 | 0.87 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr8_+_39570615 | 0.87 |
ENSDART00000142557
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr21_-_22737228 | 0.87 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr7_+_30823749 | 0.86 |
ENSDART00000085661
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr9_+_31752628 | 0.85 |
ENSDART00000060054
|
itgbl1
|
integrin, beta-like 1 |
| chr1_-_51615437 | 0.85 |
ENSDART00000152185
ENSDART00000152237 ENSDART00000129052 ENSDART00000152595 |
FBXO48
|
si:dkey-202b17.4 |
| chr7_+_25036188 | 0.84 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr24_-_8729716 | 0.84 |
ENSDART00000183624
|
tfap2a
|
transcription factor AP-2 alpha |
| chr21_-_8369073 | 0.84 |
ENSDART00000132599
|
nek6
|
NIMA-related kinase 6 |
| chr8_-_39984593 | 0.84 |
ENSDART00000140127
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr19_+_25478203 | 0.83 |
ENSDART00000132934
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr20_-_47704973 | 0.83 |
ENSDART00000174808
|
tfap2b
|
transcription factor AP-2 beta |
| chr25_+_3564220 | 0.82 |
ENSDART00000170005
|
zgc:194202
|
zgc:194202 |
| chr8_-_11229523 | 0.82 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr9_-_8454060 | 0.82 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr7_+_17229980 | 0.82 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr20_-_39735952 | 0.82 |
ENSDART00000101049
ENSDART00000137485 ENSDART00000062402 |
tpd52l1
|
tumor protein D52-like 1 |
| chr13_+_35745572 | 0.81 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr5_+_22970617 | 0.79 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr23_-_30781510 | 0.79 |
ENSDART00000183364
|
myt1a
|
myelin transcription factor 1a |
| chr7_-_44672095 | 0.78 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr8_-_18535822 | 0.78 |
ENSDART00000100558
|
nexn
|
nexilin (F actin binding protein) |
| chr4_+_8532580 | 0.78 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr18_-_41650648 | 0.78 |
ENSDART00000024087
|
fzd9b
|
frizzled class receptor 9b |
| chr24_-_8729531 | 0.78 |
ENSDART00000082346
|
tfap2a
|
transcription factor AP-2 alpha |
| chr8_+_20951590 | 0.76 |
ENSDART00000138728
|
si:dkeyp-82a1.1
|
si:dkeyp-82a1.1 |
| chr7_+_49681040 | 0.75 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr6_-_54815886 | 0.75 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr1_+_45121393 | 0.75 |
ENSDART00000142702
|
muc13a
|
mucin 13a, cell surface associated |
| chr19_-_18626515 | 0.75 |
ENSDART00000160624
|
rps18
|
ribosomal protein S18 |
| chr18_+_29145681 | 0.75 |
ENSDART00000089031
ENSDART00000193336 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr16_+_16968682 | 0.75 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr4_-_17805128 | 0.74 |
ENSDART00000128988
|
spi2
|
Spi-2 proto-oncogene |
| chr16_+_16969060 | 0.73 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr5_+_40219964 | 0.73 |
ENSDART00000114078
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr13_-_32577386 | 0.72 |
ENSDART00000016535
|
kcns3a
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3a |
| chr2_-_37896965 | 0.71 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr1_+_9290103 | 0.71 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr10_+_34394454 | 0.70 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr20_-_26420939 | 0.70 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr4_+_77810982 | 0.70 |
ENSDART00000184697
|
si:dkey-238k10.3
|
si:dkey-238k10.3 |
| chr3_+_16612574 | 0.69 |
ENSDART00000104481
|
slc17a7a
|
solute carrier family 17 (vesicular glutamate transporter), member 7a |
| chr20_+_30445971 | 0.69 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr2_+_4207209 | 0.69 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr21_+_18353703 | 0.69 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr6_-_54290227 | 0.68 |
ENSDART00000050483
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr10_+_8968203 | 0.68 |
ENSDART00000110443
ENSDART00000080772 |
fstb
|
follistatin b |
| chr15_+_58207 | 0.68 |
ENSDART00000188397
|
AXL
|
AXL receptor tyrosine kinase |
| chr9_+_34127005 | 0.67 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr4_-_40586979 | 0.67 |
ENSDART00000189361
|
si:dkey-207m2.4
|
si:dkey-207m2.4 |
| chr25_-_27541288 | 0.67 |
ENSDART00000187245
|
spam1
|
sperm adhesion molecule 1 |
| chr13_-_36535128 | 0.67 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr16_+_13855039 | 0.67 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr20_-_8419057 | 0.67 |
ENSDART00000145841
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of pgr
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.6 | 13.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.6 | 2.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.5 | 2.1 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.5 | 2.1 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.5 | 3.1 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.5 | 1.4 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.5 | 1.4 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.4 | 1.3 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.4 | 2.4 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.4 | 1.5 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.4 | 1.4 | GO:0060467 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.3 | 2.0 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.3 | 1.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.3 | 1.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 1.1 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.2 | 1.0 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.2 | 0.5 | GO:0048521 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.2 | 1.0 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.2 | 1.7 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 2.5 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.1 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 1.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.2 | 1.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.7 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.2 | 2.9 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.2 | 1.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 0.8 | GO:1904105 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 1.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.6 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 1.5 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.2 | 9.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 0.6 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 1.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 1.1 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.1 | 5.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.9 | GO:0060221 | pineal gland development(GO:0021982) retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.4 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.6 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 2.9 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.1 | 1.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 3.9 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 0.4 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.4 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 1.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.7 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.3 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.6 | GO:0032925 | regulation of activin receptor signaling pathway(GO:0032925) |
| 0.1 | 2.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.4 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.3 | GO:0050802 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) circadian sleep/wake cycle, sleep(GO:0050802) |
| 0.1 | 2.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.4 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 0.8 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 0.7 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 2.2 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 1.3 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.1 | 1.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.2 | GO:0003249 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.5 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 0.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.2 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 1.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.2 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.1 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.9 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 1.0 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 1.2 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.3 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.7 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.3 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.8 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 2.8 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.4 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.3 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 1.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 4.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 1.1 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 1.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.8 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0043435 | neutrophil homeostasis(GO:0001780) response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.8 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.3 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 4.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.7 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.8 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 1.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 3.0 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.6 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 1.6 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 1.0 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.4 | 1.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 2.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 1.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 2.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.2 | 6.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.7 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 2.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 3.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 5.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 4.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.7 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 1.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 19.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.7 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.5 | 2.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.5 | 1.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.4 | 1.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 1.1 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.4 | 3.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 1.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 0.9 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.2 | 1.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 2.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 1.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 1.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.2 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 2.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.5 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 0.6 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.2 | 1.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.0 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 1.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 2.9 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 1.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 4.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.5 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.3 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.1 | 1.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.4 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 2.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.5 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 1.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.7 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.2 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.3 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 4.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 6.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 2.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.7 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.3 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 1.0 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 10.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 5.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 14.1 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.8 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 3.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 1.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 11.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 1.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 2.9 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 2.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 3.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 2.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |