Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
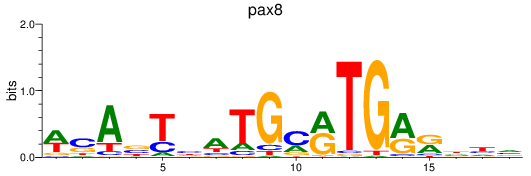
Results for pax8
Z-value: 1.36
Transcription factors associated with pax8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax8
|
ENSDARG00000015879 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax8 | dr11_v1_chr5_-_72415578_72415578 | 0.83 | 5.7e-03 | Click! |
Activity profile of pax8 motif
Sorted Z-values of pax8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23487051 | 5.45 |
ENSDART00000145496
|
icn2
|
ictacalcin 2 |
| chr4_+_25630555 | 2.21 |
ENSDART00000133425
|
acot15
|
acyl-CoA thioesterase 15 |
| chr9_+_23665777 | 1.94 |
ENSDART00000060905
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr5_+_23630384 | 1.82 |
ENSDART00000013745
|
cx39.9
|
connexin 39.9 |
| chr7_+_10610791 | 1.59 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr23_+_43255328 | 1.45 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr1_-_30457062 | 1.40 |
ENSDART00000185318
ENSDART00000157924 ENSDART00000161380 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr6_-_47840465 | 1.35 |
ENSDART00000141986
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr9_+_39027123 | 1.30 |
ENSDART00000004742
|
cps1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr23_+_26733232 | 1.29 |
ENSDART00000035080
|
zgc:158263
|
zgc:158263 |
| chr21_-_22928214 | 1.23 |
ENSDART00000182760
|
dub
|
duboraya |
| chr9_+_39027312 | 1.14 |
ENSDART00000192258
ENSDART00000164407 |
cps1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr25_-_17579701 | 1.10 |
ENSDART00000073684
|
mmp15a
|
matrix metallopeptidase 15a |
| chr7_+_19374683 | 1.09 |
ENSDART00000162700
|
snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr15_-_34892664 | 1.08 |
ENSDART00000153787
ENSDART00000099721 |
rnf183
|
ring finger protein 183 |
| chr5_-_68826177 | 1.08 |
ENSDART00000136605
|
si:ch211-283h6.4
|
si:ch211-283h6.4 |
| chr25_-_7520937 | 1.05 |
ENSDART00000170050
|
cdkn1cb
|
cyclin-dependent kinase inhibitor 1Cb |
| chr14_+_48062180 | 0.99 |
ENSDART00000056713
|
ppid
|
peptidylprolyl isomerase D |
| chr20_-_9436521 | 0.97 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr22_-_25033105 | 0.86 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr9_+_17348745 | 0.83 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr6_-_130849 | 0.74 |
ENSDART00000108710
|
LRRC8E
|
si:zfos-323e3.4 |
| chr1_+_51721851 | 0.73 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr10_+_2715548 | 0.72 |
ENSDART00000130793
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr23_+_42813415 | 0.72 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr24_-_36727922 | 0.71 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr15_+_16387088 | 0.69 |
ENSDART00000101789
|
flot2b
|
flotillin 2b |
| chr15_+_638457 | 0.67 |
ENSDART00000157162
|
znf1013
|
zinc finger protein 1013 |
| chr6_+_25215944 | 0.65 |
ENSDART00000165456
|
si:ch73-97h19.2
|
si:ch73-97h19.2 |
| chr7_+_20019125 | 0.61 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr6_+_168962 | 0.61 |
ENSDART00000182631
|
CABZ01066694.1
|
|
| chr16_+_43347966 | 0.56 |
ENSDART00000171308
|
zmp:0000000930
|
zmp:0000000930 |
| chr2_-_985417 | 0.55 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr3_+_46444625 | 0.53 |
ENSDART00000108686
|
si:ch211-66e2.3
|
si:ch211-66e2.3 |
| chr25_+_10834903 | 0.53 |
ENSDART00000155247
|
si:ch211-147g22.5
|
si:ch211-147g22.5 |
| chr19_+_46372115 | 0.53 |
ENSDART00000163935
|
med30
|
mediator complex subunit 30 |
| chr25_+_13620555 | 0.51 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr20_+_32224405 | 0.49 |
ENSDART00000062993
ENSDART00000147448 |
sesn1
|
sestrin 1 |
| chr20_+_10498986 | 0.49 |
ENSDART00000064114
|
zgc:100997
|
zgc:100997 |
| chr14_-_29906209 | 0.49 |
ENSDART00000192952
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr11_-_165288 | 0.45 |
ENSDART00000108703
ENSDART00000173151 |
tegt
|
testis enhanced gene transcript (BAX inhibitor 1) |
| chr6_-_436658 | 0.44 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr24_-_35699595 | 0.43 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr22_-_4813345 | 0.42 |
ENSDART00000190650
|
CU570881.2
|
|
| chr10_+_36037977 | 0.40 |
ENSDART00000164678
|
katnal1
|
katanin p60 subunit A-like 1 |
| chr11_-_37693019 | 0.40 |
ENSDART00000102898
|
zgc:158258
|
zgc:158258 |
| chr23_+_32039386 | 0.39 |
ENSDART00000133801
|
mylk2
|
myosin light chain kinase 2 |
| chr7_-_28549361 | 0.38 |
ENSDART00000173918
ENSDART00000054368 ENSDART00000113313 |
st5
|
suppression of tumorigenicity 5 |
| chr20_+_10498704 | 0.35 |
ENSDART00000192985
|
zgc:100997
|
zgc:100997 |
| chr16_+_46695777 | 0.35 |
ENSDART00000169767
|
rab25b
|
RAB25, member RAS oncogene family b |
| chr14_-_32491127 | 0.34 |
ENSDART00000186724
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr4_-_17803784 | 0.34 |
ENSDART00000187195
|
spi2
|
Spi-2 proto-oncogene |
| chr5_+_4298636 | 0.34 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr9_+_43797902 | 0.30 |
ENSDART00000020550
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr20_+_34605044 | 0.30 |
ENSDART00000153376
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr2_-_5942355 | 0.29 |
ENSDART00000110469
|
tmem125b
|
transmembrane protein 125b |
| chr16_+_41135018 | 0.29 |
ENSDART00000139435
|
si:ch211-53m15.2
|
si:ch211-53m15.2 |
| chr6_-_131401 | 0.29 |
ENSDART00000151251
|
LRRC8E
|
si:zfos-323e3.4 |
| chr3_+_46444999 | 0.28 |
ENSDART00000186530
|
si:ch211-66e2.3
|
si:ch211-66e2.3 |
| chr23_+_7505943 | 0.28 |
ENSDART00000135787
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr10_+_43039947 | 0.27 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr17_-_33405301 | 0.27 |
ENSDART00000157089
|
BX323819.1
|
|
| chr5_-_35888499 | 0.26 |
ENSDART00000193932
|
rxfp2l
|
relaxin/insulin-like family peptide receptor 2, like |
| chr7_+_2797795 | 0.26 |
ENSDART00000161208
|
si:ch211-217i17.1
|
si:ch211-217i17.1 |
| chr2_+_45552901 | 0.26 |
ENSDART00000131701
|
fndc7a
|
fibronectin type III domain containing 7a |
| chr4_+_36489448 | 0.24 |
ENSDART00000143181
|
znf1149
|
zinc finger protein 1149 |
| chr1_+_55239710 | 0.24 |
ENSDART00000174846
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr16_-_31435020 | 0.24 |
ENSDART00000138508
|
zgc:194210
|
zgc:194210 |
| chr22_+_5959984 | 0.23 |
ENSDART00000179378
ENSDART00000178985 ENSDART00000174635 |
BX322587.2
|
|
| chr25_-_3139805 | 0.22 |
ENSDART00000166625
|
epx
|
eosinophil peroxidase |
| chr16_+_49601838 | 0.22 |
ENSDART00000168570
ENSDART00000159236 |
si:dkey-82o10.4
|
si:dkey-82o10.4 |
| chr10_+_25726694 | 0.20 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr20_+_36234335 | 0.20 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr20_+_15164482 | 0.18 |
ENSDART00000181842
ENSDART00000063877 |
prdx6
|
peroxiredoxin 6 |
| chr5_-_12701603 | 0.17 |
ENSDART00000091124
|
aifm3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr23_+_44049509 | 0.17 |
ENSDART00000102003
|
txk
|
TXK tyrosine kinase |
| chr9_-_21825913 | 0.16 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr7_-_52388090 | 0.16 |
ENSDART00000174104
|
wdr93
|
WD repeat domain 93 |
| chr1_+_9966384 | 0.15 |
ENSDART00000132607
|
si:dkeyp-75b4.8
|
si:dkeyp-75b4.8 |
| chr10_-_24659822 | 0.15 |
ENSDART00000171842
ENSDART00000163102 |
proser1
|
proline and serine rich 1 |
| chr21_-_2162850 | 0.14 |
ENSDART00000159731
|
gb:ai877918
|
expressed sequence AI877918 |
| chr4_-_39265279 | 0.13 |
ENSDART00000164912
|
si:ch73-236c18.2
|
si:ch73-236c18.2 |
| chr7_+_3444473 | 0.13 |
ENSDART00000122334
|
si:ch211-285c6.2
|
si:ch211-285c6.2 |
| chr25_+_37285737 | 0.11 |
ENSDART00000126879
|
tmed6
|
transmembrane p24 trafficking protein 6 |
| chr3_-_16110100 | 0.10 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
| chr15_+_15314189 | 0.10 |
ENSDART00000156830
|
camkk1b
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha b |
| chr12_+_4900981 | 0.10 |
ENSDART00000171507
|
cd79b
|
CD79b molecule, immunoglobulin-associated beta |
| chr23_+_6141541 | 0.09 |
ENSDART00000105257
|
zmp:0000001139
|
zmp:0000001139 |
| chr14_+_41490262 | 0.08 |
ENSDART00000039985
|
chrnb2l
|
cholinergic receptor, nicotinic, beta 2, like |
| chr14_-_1313480 | 0.05 |
ENSDART00000097748
|
il21
|
interleukin 21 |
| chr7_-_73843720 | 0.04 |
ENSDART00000111622
|
caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr12_-_13556536 | 0.04 |
ENSDART00000166396
|
CR354395.1
|
|
| chr21_-_45891262 | 0.02 |
ENSDART00000169816
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr3_-_16110351 | 0.02 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.4 | 1.2 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.3 | 1.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 1.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.6 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.7 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.5 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 1.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.7 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 1.1 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 5.4 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.3 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 1.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.1 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 1.9 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) |
| 0.4 | 1.3 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.3 | 1.6 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 5.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 1.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.3 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 1.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.7 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 4.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |