Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
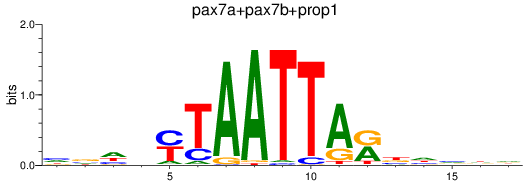
Results for pax7a+pax7b+prop1
Z-value: 1.25
Transcription factors associated with pax7a+pax7b+prop1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prop1
|
ENSDARG00000039756 | PROP paired-like homeobox 1 |
|
pax7b
|
ENSDARG00000070818 | paired box 7b |
|
pax7a
|
ENSDARG00000100398 | paired box 7a |
|
prop1
|
ENSDARG00000109950 | PROP paired-like homeobox 1 |
|
pax7a
|
ENSDARG00000111233 | paired box 7a |
|
prop1
|
ENSDARG00000114449 | PROP paired-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax7b | dr11_v1_chr23_+_20863145_20863145 | -0.90 | 1.0e-03 | Click! |
| pax7a | dr11_v1_chr11_+_41242644_41242644 | -0.82 | 6.8e-03 | Click! |
| prop1 | dr11_v1_chr13_+_8677166_8677304 | -0.75 | 2.0e-02 | Click! |
Activity profile of pax7a+pax7b+prop1 motif
Sorted Z-values of pax7a+pax7b+prop1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_34916208 | 3.43 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr10_-_34002185 | 2.03 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr2_+_6253246 | 1.93 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr8_+_45334255 | 1.72 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr16_+_47207691 | 1.60 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr17_-_4245902 | 1.60 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr22_-_20695237 | 1.54 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr9_+_8390737 | 1.47 |
ENSDART00000190891
ENSDART00000176877 ENSDART00000144851 ENSDART00000133880 ENSDART00000142233 |
zgc:113984
|
zgc:113984 |
| chr10_+_33393829 | 1.47 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr7_+_24573721 | 1.47 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr25_+_36292057 | 1.47 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr16_-_41646164 | 1.44 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr12_-_33357655 | 1.44 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr8_-_53044300 | 1.44 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr14_-_48765262 | 1.40 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr10_+_15255198 | 1.37 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr20_+_54304800 | 1.34 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr21_-_32060993 | 1.31 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr9_+_8396755 | 1.29 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr25_+_22320738 | 1.29 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr10_+_6884627 | 1.26 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr17_-_6613458 | 1.25 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr15_+_12429206 | 1.25 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr23_+_32101202 | 1.20 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr16_+_23303859 | 1.20 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr5_-_68333081 | 1.19 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr6_-_55399214 | 1.18 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr15_+_34934568 | 1.17 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr8_-_22326744 | 1.16 |
ENSDART00000137645
|
cep104
|
centrosomal protein 104 |
| chr4_+_25917915 | 1.16 |
ENSDART00000138603
|
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr1_-_40102836 | 1.14 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr5_+_37903790 | 1.14 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr1_+_35985813 | 1.13 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr19_+_1688727 | 1.13 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr17_-_40956035 | 1.11 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr23_+_42272588 | 1.10 |
ENSDART00000164907
|
CABZ01065131.1
|
|
| chr8_+_11325310 | 1.09 |
ENSDART00000142577
|
fxn
|
frataxin |
| chr14_-_14659023 | 1.07 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr19_+_2835240 | 1.06 |
ENSDART00000190838
|
CDCP1
|
CUB domain containing protein 1 |
| chr23_-_33679579 | 1.02 |
ENSDART00000188674
|
tfcp2
|
transcription factor CP2 |
| chr1_-_6085750 | 1.00 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr12_-_48188928 | 0.96 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr24_-_25166720 | 0.96 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr7_+_22313533 | 0.95 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr1_-_55068941 | 0.93 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr19_-_18127808 | 0.92 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr13_-_35808904 | 0.91 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr23_-_19225709 | 0.90 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr2_+_42871831 | 0.89 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr6_-_15491579 | 0.89 |
ENSDART00000156439
|
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr10_+_17235370 | 0.88 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr24_-_29586082 | 0.88 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr9_+_18829360 | 0.87 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr19_+_2631565 | 0.87 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr19_-_42045372 | 0.87 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr9_-_27398369 | 0.86 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr11_+_6010177 | 0.86 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr12_-_18577983 | 0.86 |
ENSDART00000193262
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr19_-_425145 | 0.85 |
ENSDART00000164905
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr12_-_4243268 | 0.85 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr18_-_26715655 | 0.85 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr18_-_15551360 | 0.85 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr15_+_29727799 | 0.84 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr17_-_21049260 | 0.84 |
ENSDART00000155300
ENSDART00000185758 |
bicc1a
|
BicC family RNA binding protein 1a |
| chr6_-_15492030 | 0.83 |
ENSDART00000156141
ENSDART00000183992 |
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr20_-_37813863 | 0.83 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr5_-_20491198 | 0.82 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr13_-_38088627 | 0.82 |
ENSDART00000175268
|
CT027676.1
|
|
| chr19_-_5103141 | 0.82 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr3_+_16722014 | 0.82 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr15_-_16155729 | 0.82 |
ENSDART00000192212
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr5_-_68779747 | 0.82 |
ENSDART00000192636
ENSDART00000188039 |
mepce
|
methylphosphate capping enzyme |
| chr11_+_12811906 | 0.82 |
ENSDART00000123445
|
rtel1
|
regulator of telomere elongation helicase 1 |
| chr25_-_18435481 | 0.81 |
ENSDART00000004771
|
poc1b
|
POC1 centriolar protein B |
| chr7_+_23515966 | 0.80 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr13_+_35474235 | 0.79 |
ENSDART00000181927
|
mkks
|
McKusick-Kaufman syndrome |
| chr21_-_2814709 | 0.79 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr6_+_21001264 | 0.77 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr22_-_19102256 | 0.77 |
ENSDART00000171866
ENSDART00000166295 |
polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr23_+_43770149 | 0.76 |
ENSDART00000024313
|
rnf150b
|
ring finger protein 150b |
| chr1_+_16625678 | 0.76 |
ENSDART00000164899
|
pcm1
|
pericentriolar material 1 |
| chr5_-_11809710 | 0.74 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr16_+_9726038 | 0.74 |
ENSDART00000192988
ENSDART00000020859 |
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr11_+_31864921 | 0.74 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr6_+_40922572 | 0.74 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr15_-_16177603 | 0.74 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr8_+_11425048 | 0.73 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr11_+_6009984 | 0.73 |
ENSDART00000185680
|
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr23_+_26026383 | 0.73 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr14_+_26247319 | 0.73 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr5_+_1109098 | 0.72 |
ENSDART00000166268
|
LO017790.1
|
|
| chr12_+_16087077 | 0.72 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr12_-_6880694 | 0.71 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr13_+_35528607 | 0.71 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr21_+_31150438 | 0.69 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr16_-_13281380 | 0.69 |
ENSDART00000103882
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr24_-_24724233 | 0.68 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr3_-_26787430 | 0.68 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr20_+_39250673 | 0.68 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr7_+_5956937 | 0.68 |
ENSDART00000170763
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr16_+_2820340 | 0.67 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr17_+_5793248 | 0.67 |
ENSDART00000153743
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr14_+_46177524 | 0.66 |
ENSDART00000074116
|
rab33ba
|
RAB33B, member RAS oncogene family a |
| chr23_+_1029450 | 0.66 |
ENSDART00000189196
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr23_+_4709607 | 0.66 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr3_-_15667713 | 0.66 |
ENSDART00000026658
|
zgc:66474
|
zgc:66474 |
| chr7_-_9873652 | 0.66 |
ENSDART00000006343
|
asb7
|
ankyrin repeat and SOCS box containing 7 |
| chr15_-_30505607 | 0.66 |
ENSDART00000155212
|
msi2b
|
musashi RNA-binding protein 2b |
| chr10_-_28118035 | 0.65 |
ENSDART00000190836
ENSDART00000088852 |
med13a
|
mediator complex subunit 13a |
| chr11_+_35171406 | 0.65 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr24_+_21514283 | 0.65 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr10_-_28117740 | 0.64 |
ENSDART00000134491
|
med13a
|
mediator complex subunit 13a |
| chr24_-_2450597 | 0.64 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr15_-_43284021 | 0.64 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr11_-_12802123 | 0.64 |
ENSDART00000104143
|
txlng
|
taxilin gamma |
| chr3_-_40658820 | 0.64 |
ENSDART00000191948
|
rnf216
|
ring finger protein 216 |
| chr2_-_42871286 | 0.63 |
ENSDART00000087823
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr8_-_7567815 | 0.63 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr8_-_39822917 | 0.63 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr5_+_6672870 | 0.63 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr10_-_31015535 | 0.62 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr24_-_34680956 | 0.62 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr15_+_19900197 | 0.61 |
ENSDART00000005221
|
thap12b
|
THAP domain containing 12b |
| chr7_+_6941583 | 0.61 |
ENSDART00000160709
ENSDART00000157634 |
rbm14b
|
RNA binding motif protein 14b |
| chr1_-_25370281 | 0.60 |
ENSDART00000050445
|
trim2a
|
tripartite motif containing 2a |
| chr21_-_27619701 | 0.60 |
ENSDART00000133441
ENSDART00000180100 |
pcnxl3
|
pecanex-like 3 (Drosophila) |
| chr15_-_2519640 | 0.60 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr22_+_35205017 | 0.60 |
ENSDART00000061315
ENSDART00000146430 |
tsc22d2
|
TSC22 domain family 2 |
| chr21_+_31150773 | 0.59 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr9_-_50000144 | 0.59 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr22_+_35275468 | 0.59 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr25_-_6432463 | 0.59 |
ENSDART00000110389
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr9_-_50001606 | 0.59 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr16_-_31622777 | 0.58 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr13_-_48161568 | 0.58 |
ENSDART00000109469
ENSDART00000188052 ENSDART00000193446 ENSDART00000189509 ENSDART00000184810 |
golga4
|
golgin A4 |
| chr20_+_29209767 | 0.58 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr7_-_69185124 | 0.57 |
ENSDART00000182217
ENSDART00000191359 |
usp10
|
ubiquitin specific peptidase 10 |
| chr20_+_29209926 | 0.57 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr4_-_2219705 | 0.57 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr15_-_17025212 | 0.57 |
ENSDART00000014465
|
hip1
|
huntingtin interacting protein 1 |
| chr17_-_48944465 | 0.56 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr5_+_28497956 | 0.56 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr22_+_35275206 | 0.56 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr25_-_27621268 | 0.56 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr16_-_28658341 | 0.56 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr23_+_28809002 | 0.55 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr9_+_48761455 | 0.55 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr2_-_3403020 | 0.55 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr6_+_40951227 | 0.55 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr13_+_49727333 | 0.54 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr17_+_52612866 | 0.54 |
ENSDART00000182828
ENSDART00000191156 ENSDART00000188814 ENSDART00000109891 |
angel1
|
angel homolog 1 (Drosophila) |
| chr16_-_34401412 | 0.54 |
ENSDART00000054020
|
hivep3b
|
human immunodeficiency virus type I enhancer binding protein 3b |
| chr13_+_2442841 | 0.54 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr23_-_14766902 | 0.54 |
ENSDART00000168113
|
gss
|
glutathione synthetase |
| chr9_-_12443726 | 0.53 |
ENSDART00000102434
|
ehhadh
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr8_+_7097929 | 0.53 |
ENSDART00000188955
ENSDART00000184772 ENSDART00000109581 |
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr8_+_18555559 | 0.53 |
ENSDART00000149523
|
tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr16_-_41787421 | 0.52 |
ENSDART00000147210
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr5_-_66397688 | 0.52 |
ENSDART00000161483
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr21_+_21195487 | 0.52 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr9_+_54039006 | 0.52 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr12_+_47081783 | 0.52 |
ENSDART00000158568
|
mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr24_-_38110779 | 0.52 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr10_-_13343831 | 0.51 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr9_-_43644261 | 0.51 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr25_+_34938317 | 0.51 |
ENSDART00000042678
|
vps4a
|
vacuolar protein sorting 4a homolog A (S. cerevisiae) |
| chr13_+_8693410 | 0.51 |
ENSDART00000138448
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr25_+_4855549 | 0.50 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr8_-_53926228 | 0.50 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr21_+_45502621 | 0.50 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr4_+_9055488 | 0.50 |
ENSDART00000155385
|
slc41a2b
|
solute carrier family 41 (magnesium transporter), member 2b |
| chr21_-_25801956 | 0.50 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr23_-_41965557 | 0.50 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr25_-_20666328 | 0.50 |
ENSDART00000098076
|
csk
|
C-terminal Src kinase |
| chr5_-_67629263 | 0.49 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr3_-_6519691 | 0.49 |
ENSDART00000165273
ENSDART00000179882 ENSDART00000172292 |
GGA3 (1 of many)
|
si:ch73-157i16.3 |
| chr22_+_11972786 | 0.49 |
ENSDART00000105788
|
mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr13_+_15838151 | 0.49 |
ENSDART00000008987
|
klc1a
|
kinesin light chain 1a |
| chr3_+_52545014 | 0.48 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr8_-_11834287 | 0.47 |
ENSDART00000187229
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr15_-_23443588 | 0.47 |
ENSDART00000170427
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr13_-_38039871 | 0.47 |
ENSDART00000140645
|
CR456624.1
|
|
| chr14_-_6286966 | 0.47 |
ENSDART00000168174
|
elp1
|
elongator complex protein 1 |
| chr18_+_39487486 | 0.47 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr20_-_29864390 | 0.47 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr25_-_37314700 | 0.46 |
ENSDART00000017807
|
igl4v8
|
immunoglobulin light 4 variable 8 |
| chr18_-_45761868 | 0.46 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr22_+_21518528 | 0.46 |
ENSDART00000148487
|
mier2
|
mesoderm induction early response 1, family member 2 |
| chr4_+_77943184 | 0.46 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr21_-_36571804 | 0.46 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr11_+_17984354 | 0.46 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr3_-_13461056 | 0.46 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr12_-_2522487 | 0.46 |
ENSDART00000022471
ENSDART00000145213 |
mapk8b
|
mitogen-activated protein kinase 8b |
| chr1_-_29139141 | 0.45 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr11_+_41540862 | 0.45 |
ENSDART00000173210
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr15_-_23442891 | 0.45 |
ENSDART00000059376
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr18_+_14619544 | 0.45 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr17_+_28533102 | 0.45 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr7_+_20383841 | 0.45 |
ENSDART00000052906
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr7_+_24645186 | 0.45 |
ENSDART00000150118
ENSDART00000150233 ENSDART00000087691 |
gba2
|
glucosidase, beta (bile acid) 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax7a+pax7b+prop1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.3 | 1.7 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.3 | 1.2 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 1.5 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.3 | 1.4 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.3 | 0.8 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.3 | 0.8 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.3 | 1.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.3 | 0.8 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 1.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 0.9 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 0.5 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.2 | 0.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 0.7 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.2 | 0.5 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 0.4 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.1 | 0.4 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.4 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.8 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 1.9 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.6 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.5 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 0.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.5 | GO:1903726 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.8 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.1 | 0.6 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.9 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 1.7 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 1.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.3 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 0.2 | GO:0044406 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 1.3 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 1.6 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.3 | GO:0014856 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 3.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.5 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.8 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.6 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 0.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 1.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.7 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.5 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.4 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 1.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.5 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 0.7 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 1.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.1 | GO:0006783 | heme biosynthetic process(GO:0006783) cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.7 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.4 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.5 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.6 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.0 | 1.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.6 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.3 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.7 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.5 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.2 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 1.1 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 1.2 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.1 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.3 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 1.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.3 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 1.4 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0003128 | heart field specification(GO:0003128) |
| 0.0 | 1.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.2 | GO:0019682 | glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.3 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.7 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.5 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.4 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.0 | GO:1901004 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.0 | 0.1 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 3.1 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.1 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 0.9 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.6 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 0.7 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 1.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.6 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.7 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 1.4 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.4 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 2.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.4 | 3.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.3 | 1.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.3 | 1.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.3 | 0.8 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.2 | 1.4 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 1.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 0.8 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 0.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 0.6 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 0.6 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 0.5 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 1.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.7 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.2 | 0.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.4 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.4 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 0.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 1.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 1.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.2 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.5 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.3 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 3.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.2 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.0 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.2 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.0 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 2.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.8 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.8 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.6 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 1.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.7 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.7 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 2.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 3.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.6 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 1.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.1 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |