Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
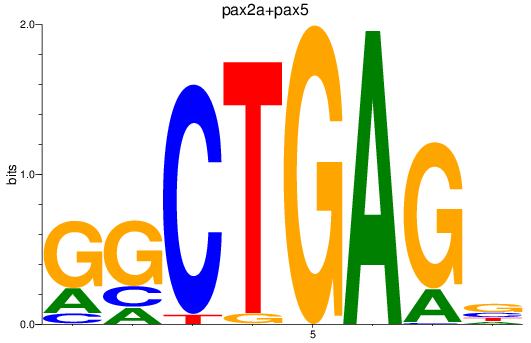
Results for pax2a+pax5
Z-value: 1.91
Transcription factors associated with pax2a+pax5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax2a
|
ENSDARG00000028148 | paired box 2a |
|
pax5
|
ENSDARG00000037383 | paired box 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax2a | dr11_v1_chr13_+_29771463_29771463 | 0.89 | 1.4e-03 | Click! |
| pax5 | dr11_v1_chr1_+_21731382_21731382 | 0.56 | 1.2e-01 | Click! |
Activity profile of pax2a+pax5 motif
Sorted Z-values of pax2a+pax5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_12031958 | 4.40 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr7_+_29952169 | 3.83 |
ENSDART00000173540
ENSDART00000173940 ENSDART00000173906 ENSDART00000173772 ENSDART00000173506 ENSDART00000039657 |
tpma
|
alpha-tropomyosin |
| chr7_+_29951997 | 3.35 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr24_-_40731305 | 3.11 |
ENSDART00000172073
|
CU633479.6
|
|
| chr15_+_32711663 | 3.04 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr7_+_29952719 | 2.96 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr3_-_55139127 | 2.89 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr3_-_55147731 | 2.58 |
ENSDART00000155871
ENSDART00000109016 ENSDART00000122904 |
hbae3
|
hemoglobin alpha embryonic-3 |
| chr3_-_55121125 | 2.44 |
ENSDART00000125092
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr24_-_40668208 | 2.32 |
ENSDART00000171543
|
smyhc1
|
slow myosin heavy chain 1 |
| chr3_-_46818001 | 2.10 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr19_-_21716593 | 2.00 |
ENSDART00000155126
|
znf516
|
zinc finger protein 516 |
| chr20_+_25340814 | 1.93 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr3_-_61205711 | 1.78 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr16_+_23978978 | 1.72 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr15_+_32711172 | 1.69 |
ENSDART00000163936
ENSDART00000168135 |
postnb
|
periostin, osteoblast specific factor b |
| chr7_+_29955368 | 1.69 |
ENSDART00000173686
|
tpma
|
alpha-tropomyosin |
| chr19_-_42551338 | 1.69 |
ENSDART00000162837
|
zgc:123103
|
zgc:123103 |
| chr9_-_22318511 | 1.65 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr25_+_31405266 | 1.65 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr3_-_46817838 | 1.65 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr13_+_17672527 | 1.64 |
ENSDART00000148269
ENSDART00000137776 |
comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr3_+_26081343 | 1.62 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr5_+_36974931 | 1.60 |
ENSDART00000193063
|
gjd1a
|
gap junction protein delta 1a |
| chr14_+_50770537 | 1.60 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr6_+_52790049 | 1.60 |
ENSDART00000002571
|
matn4
|
matrilin 4 |
| chr14_-_36378494 | 1.56 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr16_+_23921777 | 1.54 |
ENSDART00000163213
|
apoa4b.3
|
apolipoprotein A-IV b, tandem duplicate 3 |
| chr15_-_23342752 | 1.49 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr10_+_33171501 | 1.47 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr6_+_39360377 | 1.46 |
ENSDART00000028260
ENSDART00000151322 |
zgc:77517
|
zgc:77517 |
| chr21_-_27010796 | 1.46 |
ENSDART00000065398
ENSDART00000144342 ENSDART00000126542 |
ppp1r14ba
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ba |
| chr5_-_67911111 | 1.42 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr5_-_36837846 | 1.37 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr21_+_21374277 | 1.37 |
ENSDART00000079431
|
rtn2b
|
reticulon 2b |
| chr25_+_33192796 | 1.36 |
ENSDART00000125892
ENSDART00000121680 ENSDART00000014851 |
TPM1 (1 of many)
|
zgc:171719 |
| chr9_-_7539297 | 1.35 |
ENSDART00000081550
ENSDART00000081553 |
desma
|
desmin a |
| chr16_-_24518027 | 1.34 |
ENSDART00000134120
ENSDART00000143761 |
cadm4
|
cell adhesion molecule 4 |
| chr13_+_39532050 | 1.33 |
ENSDART00000019379
|
marveld1
|
MARVEL domain containing 1 |
| chr6_-_24103666 | 1.30 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr24_-_41312459 | 1.30 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr9_-_22232902 | 1.29 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr19_-_28789404 | 1.28 |
ENSDART00000191453
ENSDART00000026992 |
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr19_+_38422059 | 1.27 |
ENSDART00000035093
|
col9a2
|
procollagen, type IX, alpha 2 |
| chr11_+_3254252 | 1.26 |
ENSDART00000123568
|
pmela
|
premelanosome protein a |
| chr13_-_31452516 | 1.26 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr24_-_17047918 | 1.24 |
ENSDART00000020204
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr18_+_22302635 | 1.24 |
ENSDART00000141051
|
carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr24_+_36636208 | 1.24 |
ENSDART00000139211
|
si:ch73-334d15.4
|
si:ch73-334d15.4 |
| chr24_+_3963684 | 1.23 |
ENSDART00000182959
ENSDART00000185926 ENSDART00000167043 ENSDART00000033394 |
pfkpa
|
phosphofructokinase, platelet a |
| chr19_+_19786117 | 1.22 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr21_-_551014 | 1.22 |
ENSDART00000099252
|
vimr1
|
vimentin-related 1 |
| chr5_-_28625515 | 1.21 |
ENSDART00000190782
ENSDART00000179736 ENSDART00000131729 |
tnc
|
tenascin C |
| chr11_-_44543082 | 1.20 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr11_+_30321116 | 1.17 |
ENSDART00000187921
ENSDART00000127075 |
ugt1b1
|
UDP glucuronosyltransferase 1 family, polypeptide B1 |
| chr20_-_17041025 | 1.16 |
ENSDART00000063764
|
si:dkey-5n18.1
|
si:dkey-5n18.1 |
| chr7_+_31879986 | 1.16 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr7_-_31441420 | 1.15 |
ENSDART00000075398
|
cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr12_-_19103490 | 1.13 |
ENSDART00000060561
|
csdc2a
|
cold shock domain containing C2, RNA binding a |
| chr8_+_33035709 | 1.13 |
ENSDART00000131660
|
angptl2b
|
angiopoietin-like 2b |
| chr12_+_13256415 | 1.12 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr13_-_29424454 | 1.12 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr9_-_42989297 | 1.11 |
ENSDART00000126871
|
ttn.2
|
titin, tandem duplicate 2 |
| chr25_+_6306885 | 1.10 |
ENSDART00000142705
ENSDART00000067510 |
crabp1a
|
cellular retinoic acid binding protein 1a |
| chr5_+_38276582 | 1.09 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr4_+_5776347 | 1.09 |
ENSDART00000076494
|
ngs
|
notochord granular surface |
| chr19_-_5364649 | 1.09 |
ENSDART00000004812
|
cyt1
|
type I cytokeratin, enveloping layer |
| chr3_-_61162750 | 1.07 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr14_+_46313396 | 1.07 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr18_-_6634424 | 1.05 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr7_+_10610791 | 1.05 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr1_-_29045426 | 1.04 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr6_-_14139503 | 1.03 |
ENSDART00000089577
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr7_-_29625509 | 1.02 |
ENSDART00000173723
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr13_-_39947335 | 1.01 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr22_+_38194151 | 1.01 |
ENSDART00000121965
|
cp
|
ceruloplasmin |
| chr3_-_28258462 | 1.00 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr22_-_10459880 | 1.00 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr18_+_9171778 | 1.00 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr11_+_3254524 | 0.99 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr2_+_32780138 | 0.99 |
ENSDART00000082250
|
zgc:136930
|
zgc:136930 |
| chr3_+_32526799 | 0.99 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr1_+_16127825 | 0.98 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr21_-_28901095 | 0.98 |
ENSDART00000180820
|
cxxc5a
|
CXXC finger protein 5a |
| chr18_-_6633984 | 0.98 |
ENSDART00000185241
|
tnni1c
|
troponin I, skeletal, slow c |
| chr19_-_103289 | 0.97 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr9_+_23900703 | 0.96 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr3_-_28250722 | 0.96 |
ENSDART00000165936
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr23_-_7799184 | 0.95 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr17_+_26569601 | 0.94 |
ENSDART00000153897
|
ndnfl
|
neuron-derived neurotrophic factor , like |
| chr5_+_72108241 | 0.93 |
ENSDART00000006606
|
fabp1a
|
fatty acid binding protein 1a, liver |
| chr12_+_17865374 | 0.92 |
ENSDART00000169019
|
tmem130
|
transmembrane protein 130 |
| chr18_-_50862939 | 0.92 |
ENSDART00000180407
|
CABZ01113373.1
|
|
| chr8_-_46897734 | 0.92 |
ENSDART00000138125
|
hes2.2
|
hes family bHLH transcription factor 2, tandem duplicate 2 |
| chr6_-_18992896 | 0.91 |
ENSDART00000170228
|
sept9b
|
septin 9b |
| chr5_-_68916623 | 0.91 |
ENSDART00000141917
ENSDART00000109053 |
ank1a
|
ankyrin 1, erythrocytic a |
| chr16_-_54455573 | 0.90 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr10_+_9553935 | 0.89 |
ENSDART00000028855
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr7_+_30787903 | 0.89 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr13_+_1542493 | 0.89 |
ENSDART00000181968
|
CABZ01044281.1
|
|
| chr16_-_27628994 | 0.89 |
ENSDART00000157407
|
nacad
|
NAC alpha domain containing |
| chr16_+_32559821 | 0.88 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr18_+_17418254 | 0.87 |
ENSDART00000140191
|
ces3
|
carboxylesterase 3 |
| chr7_+_20017211 | 0.87 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr24_-_37568359 | 0.87 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr20_+_20499869 | 0.86 |
ENSDART00000036124
|
six1b
|
SIX homeobox 1b |
| chr23_+_23119008 | 0.86 |
ENSDART00000132418
|
samd11
|
sterile alpha motif domain containing 11 |
| chr3_-_46817499 | 0.86 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr19_+_1184878 | 0.86 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr5_+_43006422 | 0.85 |
ENSDART00000009182
|
aqp3a
|
aquaporin 3a |
| chr25_-_225964 | 0.85 |
ENSDART00000193424
|
CABZ01113818.1
|
|
| chr23_+_36653376 | 0.84 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr19_-_3240605 | 0.84 |
ENSDART00000105168
|
si:ch211-133n4.4
|
si:ch211-133n4.4 |
| chr16_+_5196226 | 0.83 |
ENSDART00000189704
|
soga3a
|
SOGA family member 3a |
| chr13_+_255067 | 0.82 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr5_-_55395964 | 0.82 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr7_+_44715224 | 0.82 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr12_-_4388704 | 0.81 |
ENSDART00000152168
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr3_-_12187245 | 0.81 |
ENSDART00000189553
ENSDART00000165131 |
srl
|
sarcalumenin |
| chr25_+_14017609 | 0.81 |
ENSDART00000129105
ENSDART00000125733 |
chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr19_+_56351 | 0.81 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr21_+_19445942 | 0.79 |
ENSDART00000030887
|
slc45a2
|
solute carrier family 45, member 2 |
| chr12_+_36971952 | 0.79 |
ENSDART00000125900
|
hs3st3b1b
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1b |
| chr8_-_19904124 | 0.79 |
ENSDART00000129193
|
trabd2b
|
TraB domain containing 2B |
| chr4_-_1360495 | 0.79 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr21_-_40676224 | 0.79 |
ENSDART00000162623
|
arxb
|
aristaless related homeobox b |
| chr9_-_2594410 | 0.78 |
ENSDART00000188306
ENSDART00000164276 |
sp9
|
sp9 transcription factor |
| chr18_+_402048 | 0.78 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr2_+_30916188 | 0.77 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr10_+_158590 | 0.77 |
ENSDART00000081982
|
KCNJ15
|
potassium voltage-gated channel subfamily J member 15 |
| chr1_-_5746030 | 0.76 |
ENSDART00000150863
|
nrp2a
|
neuropilin 2a |
| chr16_-_43025885 | 0.76 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr3_-_58650057 | 0.76 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr10_-_28761454 | 0.76 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr14_-_17068511 | 0.75 |
ENSDART00000163766
|
phox2bb
|
paired-like homeobox 2bb |
| chr5_-_55395384 | 0.75 |
ENSDART00000147298
ENSDART00000082577 |
prune2
|
prune homolog 2 (Drosophila) |
| chr13_-_46421682 | 0.75 |
ENSDART00000149602
|
fgfr2
|
fibroblast growth factor receptor 2 |
| chr15_+_5923851 | 0.74 |
ENSDART00000152520
ENSDART00000145827 ENSDART00000121529 |
sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr6_-_32703317 | 0.74 |
ENSDART00000064833
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr10_+_21730585 | 0.74 |
ENSDART00000188576
|
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr10_+_34394454 | 0.74 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr11_+_11201096 | 0.74 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr11_+_23957440 | 0.73 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr16_-_13730152 | 0.73 |
ENSDART00000138772
|
ttyh1
|
tweety family member 1 |
| chr17_-_32865788 | 0.73 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr21_+_40106448 | 0.73 |
ENSDART00000100166
|
serpinf1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr16_+_28596555 | 0.73 |
ENSDART00000046209
ENSDART00000141708 |
acbd7
|
acyl-CoA binding domain containing 7 |
| chr8_+_41281801 | 0.72 |
ENSDART00000084439
|
olfml2a
|
olfactomedin-like 2A |
| chr13_+_4405282 | 0.72 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr9_+_307863 | 0.72 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr2_-_9646857 | 0.72 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr8_-_21268303 | 0.71 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr11_-_18253111 | 0.71 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr21_+_5129513 | 0.70 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr18_-_46010 | 0.70 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr8_-_1051438 | 0.70 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr1_-_31534089 | 0.69 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr10_-_17103651 | 0.69 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr3_-_41791178 | 0.69 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr11_-_35763323 | 0.69 |
ENSDART00000026017
|
bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr21_-_28920245 | 0.69 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr4_+_16323970 | 0.69 |
ENSDART00000190651
|
BX322608.1
|
|
| chr9_+_54679221 | 0.68 |
ENSDART00000167769
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr5_+_43603794 | 0.68 |
ENSDART00000134633
|
si:dkey-40c11.2
|
si:dkey-40c11.2 |
| chr10_-_26729930 | 0.68 |
ENSDART00000145532
|
fgf13b
|
fibroblast growth factor 13b |
| chr21_-_23746916 | 0.68 |
ENSDART00000017229
|
ncam1a
|
neural cell adhesion molecule 1a |
| chr16_-_13921589 | 0.68 |
ENSDART00000023543
|
rcvrn2
|
recoverin 2 |
| chr19_+_31771270 | 0.68 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr18_-_48530221 | 0.68 |
ENSDART00000188134
ENSDART00000142107 |
kcnj1a.2
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 2 |
| chr8_+_39634114 | 0.67 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr12_+_47909026 | 0.67 |
ENSDART00000192472
|
tbata
|
thymus, brain and testes associated |
| chr9_-_7683799 | 0.67 |
ENSDART00000102713
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr10_-_22803740 | 0.67 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr23_-_4091009 | 0.67 |
ENSDART00000109807
|
FQ323119.1
|
|
| chr14_+_20893065 | 0.66 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr4_-_17055782 | 0.66 |
ENSDART00000134595
|
sox5
|
SRY (sex determining region Y)-box 5 |
| chr14_+_33722950 | 0.66 |
ENSDART00000075312
|
apln
|
apelin |
| chr12_+_45676667 | 0.66 |
ENSDART00000016553
|
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr16_-_28856112 | 0.66 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr5_-_19394440 | 0.65 |
ENSDART00000163771
|
foxn4
|
forkhead box N4 |
| chr23_+_24705424 | 0.65 |
ENSDART00000104029
|
c1qtnf12
|
C1q and TNF related 12 |
| chr8_-_31075015 | 0.65 |
ENSDART00000010993
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr14_-_21219659 | 0.65 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr7_-_30177691 | 0.65 |
ENSDART00000046689
|
tmed3
|
transmembrane p24 trafficking protein 3 |
| chr24_-_2947393 | 0.65 |
ENSDART00000166661
ENSDART00000147110 |
tubb6
|
tubulin, beta 6 class V |
| chr17_-_26911852 | 0.64 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr6_-_13187168 | 0.64 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr2_+_21982911 | 0.64 |
ENSDART00000190722
ENSDART00000044371 ENSDART00000134912 |
tox
|
thymocyte selection-associated high mobility group box |
| chr5_+_65991152 | 0.64 |
ENSDART00000097756
|
lcn15
|
lipocalin 15 |
| chr9_+_23665777 | 0.64 |
ENSDART00000060905
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr9_-_18877597 | 0.64 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr9_-_7684002 | 0.64 |
ENSDART00000016360
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr5_+_27432958 | 0.63 |
ENSDART00000124705
|
histh1l
|
histone H1 like |
| chr16_+_29043813 | 0.63 |
ENSDART00000122681
|
nes
|
nestin |
| chr5_+_34407763 | 0.63 |
ENSDART00000188849
ENSDART00000145127 |
lamc3
|
laminin, gamma 3 |
| chr16_-_29277164 | 0.62 |
ENSDART00000058870
|
rhbg
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr9_+_34641237 | 0.62 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr14_+_32022272 | 0.62 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr21_-_10773344 | 0.62 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr8_-_23416362 | 0.62 |
ENSDART00000063005
|
gpr173
|
G protein-coupled receptor 173 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax2a+pax5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0097435 | fibril organization(GO:0097435) |
| 0.5 | 0.5 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.4 | 1.3 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.4 | 1.6 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.4 | 1.2 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.4 | 1.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.4 | 1.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.4 | 0.7 | GO:0014808 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.3 | 1.0 | GO:1903792 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.3 | 1.4 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.3 | 2.3 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 1.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 1.2 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.3 | 0.9 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.3 | 0.9 | GO:0097623 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.3 | 0.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.3 | 1.0 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.2 | 1.0 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 4.4 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.2 | 0.7 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 1.0 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.2 | 5.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 1.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 0.7 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.2 | 1.3 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.2 | 0.7 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 1.1 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.2 | 0.6 | GO:0019557 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 1.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 0.6 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 0.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 1.0 | GO:0055016 | hypochord development(GO:0055016) |
| 0.2 | 1.8 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 2.1 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.2 | 0.6 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.2 | 0.8 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.2 | 0.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 1.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 6.6 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 1.6 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 2.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 0.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.7 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.4 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.1 | 16.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 0.9 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.6 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.1 | 0.7 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.4 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.1 | 0.5 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 0.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.7 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.7 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.5 | GO:0051503 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.5 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 1.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.7 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 0.3 | GO:1902746 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 1.4 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.4 | GO:0061032 | pericardium development(GO:0060039) visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.5 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.2 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.5 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.1 | 0.3 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.8 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.4 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.1 | GO:0060035 | notochord cell development(GO:0060035) |
| 0.1 | 1.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.4 | GO:2000561 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 1.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.3 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.3 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.4 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.6 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 1.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.7 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.1 | GO:0061217 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.5 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 1.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.1 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.1 | 1.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.6 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.2 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.1 | 0.2 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.4 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.4 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.3 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 0.8 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.8 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.6 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.0 | 0.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.5 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.1 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.0 | 1.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.3 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.8 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 1.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.9 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.5 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 1.1 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.7 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 4.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0048890 | lateral line ganglion development(GO:0048890) posterior lateral line ganglion development(GO:0048917) |
| 0.0 | 0.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 2.9 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.3 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.0 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.4 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0015893 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.9 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.7 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0007603 | phototransduction, visible light(GO:0007603) light absorption(GO:0016037) |
| 0.0 | 0.8 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 2.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.8 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.6 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.4 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.7 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.2 | GO:0051893 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.5 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.4 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.4 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.3 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.7 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.4 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 1.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.4 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.7 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.4 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.2 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.2 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.0 | 0.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.3 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.3 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 0.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.5 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.2 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.5 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.3 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.0 | 1.5 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 3.9 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 1.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.7 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.5 | GO:0051588 | regulation of neurotransmitter transport(GO:0051588) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0098926 | postsynaptic signal transduction(GO:0098926) |
| 0.0 | 0.0 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.7 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 2.9 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.0 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.6 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.0 | GO:0018158 | protein oxidation(GO:0018158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 1.6 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 8.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 0.6 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 3.2 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 1.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.7 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 12.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.7 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 8.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 2.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.9 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 2.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.2 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 3.5 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 3.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 6.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.1 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.9 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 1.3 | GO:0034704 | voltage-gated calcium channel complex(GO:0005891) calcium channel complex(GO:0034704) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.9 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 14.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.0 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.5 | 1.6 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.4 | 5.0 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.3 | 1.0 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.3 | 1.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.3 | 0.9 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.3 | 1.1 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.3 | 1.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.7 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.8 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.2 | 0.7 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 3.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 1.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.6 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 1.6 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 1.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.9 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 1.4 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.4 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.3 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 1.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.3 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.6 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 9.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 1.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.2 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 4.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.3 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 1.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.3 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.6 | GO:0015183 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 1.3 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 1.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 1.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 6.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 1.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.0 | 0.6 | GO:0051371 | actinin binding(GO:0042805) muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0098918 | structural constituent of presynaptic active zone(GO:0098882) structural constituent of synapse(GO:0098918) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.3 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.0 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 9.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 4.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 2.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.4 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.1 | 1.0 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.4 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.1 | 0.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |