Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
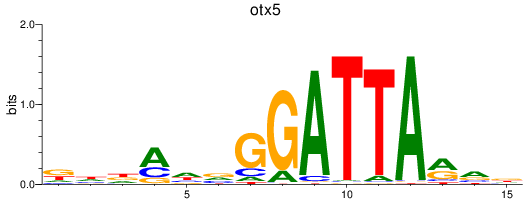
Results for otx5
Z-value: 0.87
Transcription factors associated with otx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
otx5
|
ENSDARG00000043483 | orthodenticle homolog 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx5 | dr11_v1_chr15_+_47903864_47903864 | -0.72 | 2.9e-02 | Click! |
Activity profile of otx5 motif
Sorted Z-values of otx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_25140904 | 1.17 |
ENSDART00000143114
|
si:dkeyp-20e4.8
|
si:dkeyp-20e4.8 |
| chr4_-_77125693 | 1.14 |
ENSDART00000174256
|
CU467646.3
|
|
| chr12_-_4301234 | 1.07 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr4_-_77130289 | 1.00 |
ENSDART00000174380
|
CU467646.7
|
|
| chr5_-_3584815 | 0.73 |
ENSDART00000152108
|
hspb1
|
heat shock protein, alpha-crystallin-related, 1 |
| chr10_-_1961930 | 0.69 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr10_-_1961576 | 0.68 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr16_+_40043673 | 0.57 |
ENSDART00000102552
ENSDART00000125484 |
trmt11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr12_+_3912544 | 0.56 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr16_+_52343905 | 0.55 |
ENSDART00000131051
|
ifnlr1
|
interferon lambda receptor 1 |
| chr4_-_8030583 | 0.53 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr14_-_41308561 | 0.52 |
ENSDART00000138232
|
arl13a
|
ADP-ribosylation factor-like 13A |
| chr11_+_18037729 | 0.51 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr8_+_36560019 | 0.51 |
ENSDART00000136418
ENSDART00000061378 ENSDART00000185237 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr23_-_45955177 | 0.49 |
ENSDART00000165963
ENSDART00000186649 ENSDART00000185773 |
LO017850.1
|
|
| chr22_+_25582707 | 0.49 |
ENSDART00000184440
|
aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr24_+_36317544 | 0.47 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr9_+_18829360 | 0.46 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr3_-_5644028 | 0.44 |
ENSDART00000019957
|
ddx39ab
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Ab |
| chr4_-_77432218 | 0.44 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr24_+_10898671 | 0.41 |
ENSDART00000106272
|
si:dkey-37o8.1
|
si:dkey-37o8.1 |
| chr5_+_26847190 | 0.39 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr22_-_26289549 | 0.39 |
ENSDART00000043774
|
sde2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr11_+_3499111 | 0.38 |
ENSDART00000113096
ENSDART00000171621 |
pusl1
|
pseudouridylate synthase-like 1 |
| chr18_+_7639401 | 0.38 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr19_-_2115040 | 0.37 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr8_+_50150834 | 0.37 |
ENSDART00000056074
|
entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr17_-_8592824 | 0.37 |
ENSDART00000127022
|
CU462878.1
|
|
| chr9_+_3519191 | 0.36 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr17_+_16046314 | 0.36 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr17_+_35097024 | 0.36 |
ENSDART00000026152
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr11_+_29671661 | 0.36 |
ENSDART00000024318
ENSDART00000165024 |
rnf207a
|
ring finger protein 207a |
| chr5_-_57652168 | 0.35 |
ENSDART00000166245
|
gb:eh456644
|
expressed sequence EH456644 |
| chr17_+_16046132 | 0.35 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr23_+_39963599 | 0.34 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr20_+_30725778 | 0.34 |
ENSDART00000062532
|
nhsl1b
|
NHS-like 1b |
| chr22_-_26484268 | 0.33 |
ENSDART00000110293
|
zgc:194551
|
zgc:194551 |
| chr5_-_4297459 | 0.33 |
ENSDART00000018895
|
srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr18_-_38245062 | 0.32 |
ENSDART00000189092
|
nat10
|
N-acetyltransferase 10 |
| chr15_-_6976851 | 0.32 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr8_+_2543525 | 0.31 |
ENSDART00000129569
ENSDART00000128227 |
gle1
|
GLE1 RNA export mediator homolog (yeast) |
| chr2_-_31686353 | 0.31 |
ENSDART00000126177
ENSDART00000056679 |
e2f5
|
E2F transcription factor 5 |
| chr14_+_1124409 | 0.30 |
ENSDART00000190043
ENSDART00000106708 ENSDART00000160246 |
sec24b
|
SEC24 homolog B, COPII coat complex component |
| chr6_+_40832635 | 0.30 |
ENSDART00000011931
|
ruvbl1
|
RuvB-like AAA ATPase 1 |
| chr16_-_25741225 | 0.30 |
ENSDART00000130641
|
bcl3
|
B cell CLL/lymphoma 3 |
| chr8_+_21297217 | 0.30 |
ENSDART00000142836
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr24_+_19518303 | 0.29 |
ENSDART00000027022
ENSDART00000056080 |
sulf1
|
sulfatase 1 |
| chr8_+_25959940 | 0.29 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr1_-_30762264 | 0.28 |
ENSDART00000085454
|
dis3
|
DIS3 exosome endoribonuclease and 3'-5' exoribonuclease |
| chr3_+_3610140 | 0.27 |
ENSDART00000104977
|
pick1
|
protein interacting with prkca 1 |
| chr4_+_52177103 | 0.27 |
ENSDART00000166189
|
si:dkey-16b10.1
|
si:dkey-16b10.1 |
| chr12_-_30777540 | 0.26 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr3_-_49138004 | 0.26 |
ENSDART00000167173
|
gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr16_-_48428035 | 0.26 |
ENSDART00000190554
|
rad21a
|
RAD21 cohesin complex component a |
| chr20_-_33566640 | 0.26 |
ENSDART00000159729
|
si:dkey-65b13.9
|
si:dkey-65b13.9 |
| chr4_-_14470071 | 0.26 |
ENSDART00000143773
|
plxnb2a
|
plexin b2a |
| chr25_-_17590971 | 0.26 |
ENSDART00000189942
|
mmp15a
|
matrix metallopeptidase 15a |
| chr22_-_12726556 | 0.25 |
ENSDART00000105776
|
fam207a
|
family with sequence similarity 207, member A |
| chr16_+_10557504 | 0.25 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr25_-_34740627 | 0.25 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr22_-_16154771 | 0.25 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr8_-_50979047 | 0.24 |
ENSDART00000184788
ENSDART00000180906 |
zgc:91909
|
zgc:91909 |
| chr24_-_37484123 | 0.24 |
ENSDART00000111623
|
cluap1
|
clusterin associated protein 1 |
| chr10_-_17550239 | 0.24 |
ENSDART00000057513
|
ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr5_+_31791001 | 0.24 |
ENSDART00000043010
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr7_-_59515569 | 0.24 |
ENSDART00000163343
ENSDART00000165457 ENSDART00000163745 |
slx1b
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr3_+_9071806 | 0.24 |
ENSDART00000162372
|
znf985
|
zinc finger protein 985 |
| chr20_+_34671386 | 0.23 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr24_+_17005647 | 0.23 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr5_+_4016271 | 0.23 |
ENSDART00000113627
ENSDART00000105832 ENSDART00000121415 |
ggnbp2
|
gametogenetin binding protein 2 |
| chr20_-_28931901 | 0.23 |
ENSDART00000153082
ENSDART00000046042 |
susd6
|
sushi domain containing 6 |
| chr12_-_30777743 | 0.22 |
ENSDART00000148888
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr5_-_69180227 | 0.22 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr6_-_39521832 | 0.22 |
ENSDART00000065038
|
atf1
|
activating transcription factor 1 |
| chr4_-_9764767 | 0.22 |
ENSDART00000164328
ENSDART00000147699 |
mical3b
|
microtubule associated monooxygenase, calponin and LIM domain containing 3b |
| chr19_+_2631565 | 0.22 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr5_-_30074332 | 0.22 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr3_-_27647845 | 0.22 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr17_+_50701748 | 0.22 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr4_-_43919359 | 0.21 |
ENSDART00000184163
ENSDART00000158019 ENSDART00000191745 ENSDART00000181638 ENSDART00000150827 |
znf1104
|
zinc finger protein 1104 |
| chr15_-_15469079 | 0.21 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr4_-_38608436 | 0.21 |
ENSDART00000191971
|
BX324212.2
|
|
| chr17_-_51893123 | 0.20 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr12_+_17042754 | 0.20 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr19_+_3215466 | 0.20 |
ENSDART00000181288
|
zgc:86598
|
zgc:86598 |
| chr3_+_35608385 | 0.20 |
ENSDART00000193219
ENSDART00000132703 |
traf7
|
TNF receptor-associated factor 7 |
| chr14_+_3038473 | 0.20 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr3_+_16841942 | 0.20 |
ENSDART00000023985
ENSDART00000145317 |
stk17al
|
serine/threonine kinase 17a like |
| chr1_+_45217425 | 0.20 |
ENSDART00000179983
ENSDART00000074683 |
EVI5L
|
si:ch211-239f4.1 |
| chr17_+_25426091 | 0.20 |
ENSDART00000149241
ENSDART00000121848 |
srrm1
|
serine/arginine repetitive matrix 1 |
| chr1_-_11075403 | 0.20 |
ENSDART00000102903
ENSDART00000170290 |
dmd
|
dystrophin |
| chr10_+_6013076 | 0.20 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr20_+_6533260 | 0.20 |
ENSDART00000135005
ENSDART00000166356 |
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr23_+_39854566 | 0.20 |
ENSDART00000190423
ENSDART00000164473 ENSDART00000161881 |
si:ch73-217b7.1
|
si:ch73-217b7.1 |
| chr4_-_13614797 | 0.20 |
ENSDART00000138366
ENSDART00000165212 |
irf5
|
interferon regulatory factor 5 |
| chr3_-_32170850 | 0.20 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr13_+_1015749 | 0.19 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr4_+_64756094 | 0.19 |
ENSDART00000150856
ENSDART00000188974 ENSDART00000183321 ENSDART00000188031 ENSDART00000168077 ENSDART00000157953 ENSDART00000193880 ENSDART00000167975 |
znf1018
|
zinc finger protein 1018 |
| chr4_+_34860183 | 0.19 |
ENSDART00000123281
ENSDART00000192960 ENSDART00000140637 ENSDART00000192576 ENSDART00000188148 |
znf985
|
zinc finger protein 985 |
| chr4_+_51339670 | 0.19 |
ENSDART00000126791
ENSDART00000179695 ENSDART00000162443 ENSDART00000167799 ENSDART00000170092 ENSDART00000181068 ENSDART00000183460 |
si:dkeyp-82b4.3
|
si:dkeyp-82b4.3 |
| chr5_+_24287927 | 0.19 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr1_-_46859398 | 0.19 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr4_-_75651505 | 0.18 |
ENSDART00000115143
|
si:dkey-71l4.2
|
si:dkey-71l4.2 |
| chr16_-_40043322 | 0.18 |
ENSDART00000145278
|
si:dkey-29b11.3
|
si:dkey-29b11.3 |
| chr16_+_25285998 | 0.18 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr2_+_7192966 | 0.18 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr16_+_28547157 | 0.18 |
ENSDART00000109450
ENSDART00000165687 |
fam171a1
|
family with sequence similarity 171, member A1 |
| chr21_-_43428040 | 0.18 |
ENSDART00000148325
|
stk26
|
serine/threonine protein kinase 26 |
| chr15_+_40079468 | 0.18 |
ENSDART00000154947
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr12_+_19138452 | 0.17 |
ENSDART00000141346
ENSDART00000066397 |
phf5a
|
PHD finger protein 5A |
| chr4_+_18441988 | 0.17 |
ENSDART00000040827
|
ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr18_+_45862414 | 0.17 |
ENSDART00000024615
|
rnpepl1
|
arginyl aminopeptidase like 1 |
| chr18_+_6641542 | 0.17 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr20_+_23742574 | 0.17 |
ENSDART00000050559
|
sh3rf1
|
SH3 domain containing ring finger 1 |
| chr5_-_69180587 | 0.17 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr4_+_42175261 | 0.17 |
ENSDART00000162193
|
si:ch211-142b24.2
|
si:ch211-142b24.2 |
| chr9_-_54981471 | 0.17 |
ENSDART00000097318
|
fancb
|
Fanconi anemia, complementation group B |
| chr6_+_32834760 | 0.17 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr10_-_44467384 | 0.17 |
ENSDART00000186382
|
CABZ01072089.1
|
|
| chr16_-_21692024 | 0.16 |
ENSDART00000123597
|
si:ch211-154o6.2
|
si:ch211-154o6.2 |
| chr8_+_23391263 | 0.16 |
ENSDART00000149687
|
si:dkey-16n15.6
|
si:dkey-16n15.6 |
| chr4_+_45610254 | 0.16 |
ENSDART00000150866
|
znf1038
|
zinc finger protein 1038 |
| chr21_-_22117085 | 0.16 |
ENSDART00000146673
|
slc35f2
|
solute carrier family 35, member F2 |
| chr4_+_69712223 | 0.16 |
ENSDART00000163035
ENSDART00000164475 |
si:dkey-238o14.1
zgc:174944
|
si:dkey-238o14.1 zgc:174944 |
| chr4_+_61865790 | 0.15 |
ENSDART00000166793
|
si:dkey-146c18.5
|
si:dkey-146c18.5 |
| chr4_-_43185261 | 0.15 |
ENSDART00000150380
|
si:dkey-16p6.2
|
si:dkey-16p6.2 |
| chr2_-_4032732 | 0.15 |
ENSDART00000158335
|
rab18b
|
RAB18B, member RAS oncogene family |
| chr8_+_2656231 | 0.15 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr10_-_26218354 | 0.15 |
ENSDART00000180764
|
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr4_+_58667348 | 0.15 |
ENSDART00000186596
ENSDART00000180673 |
CR388148.2
|
|
| chr4_+_69375564 | 0.14 |
ENSDART00000150334
|
si:dkey-246j6.1
|
si:dkey-246j6.1 |
| chr17_-_25737452 | 0.14 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr12_+_11352630 | 0.14 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr15_+_5973909 | 0.14 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr13_-_35527697 | 0.14 |
ENSDART00000016793
|
memo1
|
mediator of cell motility 1 |
| chr6_-_15604417 | 0.14 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr19_-_32518556 | 0.14 |
ENSDART00000103410
|
zbtb8b
|
zinc finger and BTB domain containing 8B |
| chr4_+_65271779 | 0.14 |
ENSDART00000134257
|
znf1027
|
zinc finger protein 1027 |
| chr11_+_6882362 | 0.14 |
ENSDART00000144181
|
klhl26
|
kelch-like family member 26 |
| chr7_-_17779644 | 0.13 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr20_+_29587995 | 0.13 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr4_+_43225728 | 0.13 |
ENSDART00000158639
ENSDART00000141339 ENSDART00000137770 ENSDART00000166019 |
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr9_-_3519253 | 0.13 |
ENSDART00000169586
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr7_-_65114067 | 0.13 |
ENSDART00000110614
ENSDART00000098277 |
tmem41b
|
transmembrane protein 41B |
| chr6_+_45347219 | 0.13 |
ENSDART00000188240
|
LO017951.1
|
|
| chr8_+_23390844 | 0.13 |
ENSDART00000184556
|
si:dkey-16n15.6
|
si:dkey-16n15.6 |
| chr3_+_25907266 | 0.13 |
ENSDART00000170324
ENSDART00000192633 |
tom1
|
target of myb1 membrane trafficking protein |
| chr4_+_49664256 | 0.13 |
ENSDART00000155251
|
znf974
|
zinc finger protein 974 |
| chr4_+_33818711 | 0.13 |
ENSDART00000186452
|
znf1026
|
zinc finger protein 1026 |
| chr17_-_7371564 | 0.13 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr11_-_11336986 | 0.13 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr8_+_17168114 | 0.13 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr1_+_26626824 | 0.13 |
ENSDART00000158193
|
coro2a
|
coronin, actin binding protein, 2A |
| chr8_+_23355484 | 0.12 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr19_+_42694211 | 0.12 |
ENSDART00000180153
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr17_-_42492668 | 0.12 |
ENSDART00000183946
|
CR925755.2
|
|
| chr3_-_58165254 | 0.12 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr4_-_50722304 | 0.12 |
ENSDART00000150480
|
znf1110
|
zinc finger protein 1110 |
| chr6_-_17999776 | 0.12 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr8_-_48704688 | 0.12 |
ENSDART00000165307
|
pimr182
|
Pim proto-oncogene, serine/threonine kinase, related 182 |
| chr3_+_12725343 | 0.12 |
ENSDART00000188583
|
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr17_+_10748366 | 0.12 |
ENSDART00000018683
ENSDART00000097274 |
ATG14
|
zgc:113944 |
| chr6_+_46481024 | 0.12 |
ENSDART00000155596
|
si:dkey-7k24.5
|
si:dkey-7k24.5 |
| chr19_-_9503473 | 0.11 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr25_-_17552299 | 0.11 |
ENSDART00000154327
|
si:dkey-44k1.5
|
si:dkey-44k1.5 |
| chr11_+_25583950 | 0.11 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr1_+_41690402 | 0.11 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr13_-_14269626 | 0.11 |
ENSDART00000079176
|
gfra4a
|
GDNF family receptor alpha 4a |
| chr4_+_43245188 | 0.11 |
ENSDART00000150310
|
si:dkey-29j8.1
|
si:dkey-29j8.1 |
| chr11_+_11267829 | 0.11 |
ENSDART00000026814
ENSDART00000173346 ENSDART00000151926 |
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr23_-_40250886 | 0.11 |
ENSDART00000041912
|
tgm1l3
|
transglutaminase 1 like 3 |
| chr4_+_58661331 | 0.11 |
ENSDART00000188763
|
CR388148.2
|
|
| chr11_+_31680513 | 0.11 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr6_-_59381391 | 0.10 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr17_-_8656155 | 0.10 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr8_-_5850472 | 0.10 |
ENSDART00000179217
|
CABZ01102147.1
|
|
| chr8_+_54263946 | 0.10 |
ENSDART00000193119
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr7_-_12789251 | 0.10 |
ENSDART00000052750
|
adamtsl3
|
ADAMTS-like 3 |
| chr23_+_27352072 | 0.09 |
ENSDART00000144762
|
si:dkey-9p24.5
|
si:dkey-9p24.5 |
| chr4_+_65487457 | 0.09 |
ENSDART00000162017
ENSDART00000193241 |
si:dkeyp-5g9.1
|
si:dkeyp-5g9.1 |
| chr10_-_5016997 | 0.09 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr23_+_44349252 | 0.09 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr18_+_33490315 | 0.09 |
ENSDART00000140885
ENSDART00000155210 |
v2rh1
|
vomeronasal 2 receptor, h1 |
| chr4_+_63992019 | 0.09 |
ENSDART00000158645
|
CR790365.2
|
|
| chr20_-_34754617 | 0.08 |
ENSDART00000148066
|
znf395b
|
zinc finger protein 395b |
| chr10_-_8294965 | 0.08 |
ENSDART00000167380
|
plpp1a
|
phospholipid phosphatase 1a |
| chr1_-_29761086 | 0.08 |
ENSDART00000136760
|
alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr4_+_31450683 | 0.08 |
ENSDART00000169303
|
znf996
|
zinc finger protein 996 |
| chr4_+_33012407 | 0.08 |
ENSDART00000151873
|
si:dkey-26h11.2
|
si:dkey-26h11.2 |
| chr4_-_31795106 | 0.08 |
ENSDART00000164175
|
si:dkey-19c16.12
|
si:dkey-19c16.12 |
| chr15_-_36727462 | 0.08 |
ENSDART00000085971
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr2_+_47489679 | 0.08 |
ENSDART00000074968
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr13_+_30169681 | 0.08 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr4_-_63769637 | 0.07 |
ENSDART00000158757
|
znf1098
|
zinc finger protein 1098 |
| chr7_-_17780048 | 0.07 |
ENSDART00000183336
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr9_+_1313418 | 0.07 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr15_+_31426768 | 0.07 |
ENSDART00000014270
|
or103-1
|
odorant receptor, family C, subfamily 103, member 1 |
| chr4_-_61419027 | 0.07 |
ENSDART00000181567
ENSDART00000187397 ENSDART00000180671 ENSDART00000164053 ENSDART00000187395 |
znf1021
|
zinc finger protein 1021 |
| chr4_-_71214516 | 0.07 |
ENSDART00000169997
|
si:ch211-205a14.7
|
si:ch211-205a14.7 |
| chr4_+_60428812 | 0.07 |
ENSDART00000162049
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of otx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.2 | 0.7 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.2 | 0.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 0.5 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.9 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.4 | GO:0010664 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.1 | 0.4 | GO:0046048 | ribonucleoside diphosphate catabolic process(GO:0009191) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 0.5 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.6 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.2 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.2 | GO:0010935 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.2 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 1.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.3 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.3 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.3 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) regulation of heart growth(GO:0060420) |
| 0.0 | 0.3 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.2 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 0.2 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.3 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.0 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.3 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.5 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.2 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.3 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.0 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.6 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 1.4 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.9 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.0 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.1 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |