Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
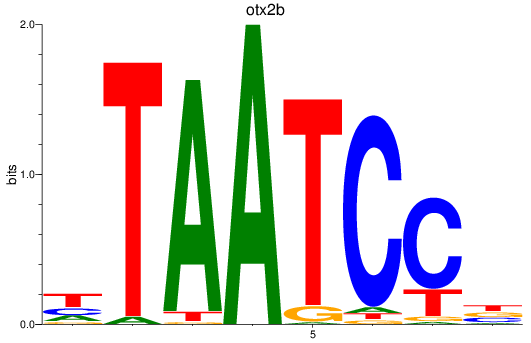
Results for otx2b
Z-value: 2.20
Transcription factors associated with otx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
otx2b
|
ENSDARG00000011235 | orthodenticle homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx2b | dr11_v1_chr17_-_44249720_44249720 | 0.78 | 1.3e-02 | Click! |
Activity profile of otx2b motif
Sorted Z-values of otx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_26144765 | 10.88 |
ENSDART00000146267
ENSDART00000043932 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr15_+_32727848 | 5.32 |
ENSDART00000161361
|
postnb
|
periostin, osteoblast specific factor b |
| chr5_-_28625515 | 4.26 |
ENSDART00000190782
ENSDART00000179736 ENSDART00000131729 |
tnc
|
tenascin C |
| chr23_+_36063599 | 3.74 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr14_-_4273396 | 3.54 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr8_-_54077740 | 3.45 |
ENSDART00000027000
|
rho
|
rhodopsin |
| chr22_+_5103349 | 3.42 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr14_+_46313135 | 3.41 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr7_-_13882988 | 3.34 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr14_-_36378494 | 3.33 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr15_+_32711172 | 3.25 |
ENSDART00000163936
ENSDART00000168135 |
postnb
|
periostin, osteoblast specific factor b |
| chr15_+_33070939 | 3.17 |
ENSDART00000164928
|
mab21l1
|
mab-21-like 1 |
| chr23_-_6641223 | 3.14 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr1_+_17593392 | 2.95 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
| chr16_+_32059785 | 2.86 |
ENSDART00000134459
|
si:dkey-40m6.8
|
si:dkey-40m6.8 |
| chr5_-_28606916 | 2.67 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr7_-_41489822 | 2.60 |
ENSDART00000006724
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr7_+_67467702 | 2.51 |
ENSDART00000168460
ENSDART00000170322 |
rpl13
|
ribosomal protein L13 |
| chr8_+_6954984 | 2.51 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr13_-_39947335 | 2.16 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr3_+_23703704 | 2.13 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr15_-_2652640 | 2.11 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr9_+_26103814 | 2.10 |
ENSDART00000026011
|
efnb2a
|
ephrin-B2a |
| chr16_+_20926673 | 2.08 |
ENSDART00000009827
|
hoxa2b
|
homeobox A2b |
| chr3_+_1492174 | 2.07 |
ENSDART00000112979
|
sox10
|
SRY (sex determining region Y)-box 10 |
| chr11_+_2172335 | 2.00 |
ENSDART00000170593
|
hoxc12b
|
homeobox C12b |
| chr7_-_41489997 | 1.97 |
ENSDART00000174300
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr10_+_32104305 | 1.93 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr7_+_71335815 | 1.91 |
ENSDART00000102756
|
sod3a
|
superoxide dismutase 3, extracellular a |
| chr6_-_28980756 | 1.89 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr9_+_7724152 | 1.87 |
ENSDART00000061716
|
mnx2a
|
motor neuron and pancreas homeobox 2a |
| chr19_-_8880688 | 1.86 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr21_-_20341836 | 1.84 |
ENSDART00000176689
|
rbp4l
|
retinol binding protein 4, like |
| chr3_-_28828242 | 1.78 |
ENSDART00000151445
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr16_+_42830152 | 1.71 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr8_-_22964486 | 1.69 |
ENSDART00000112381
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr20_+_53441935 | 1.66 |
ENSDART00000175214
|
apobb.2
|
apolipoprotein Bb, tandem duplicate 2 |
| chr6_-_20875111 | 1.65 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr16_-_33059246 | 1.65 |
ENSDART00000171718
ENSDART00000168305 ENSDART00000166401 |
snap91
|
synaptosomal-associated protein 91 |
| chr11_-_15090564 | 1.63 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr3_+_23743139 | 1.59 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr10_+_4095917 | 1.58 |
ENSDART00000114533
|
mn1a
|
meningioma 1a |
| chr3_+_23742868 | 1.54 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr21_-_20342096 | 1.51 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr17_-_12389259 | 1.51 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr5_+_22510639 | 1.48 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr18_-_22753637 | 1.48 |
ENSDART00000181589
ENSDART00000009912 |
hsf4
|
heat shock transcription factor 4 |
| chr16_+_42829735 | 1.47 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr5_-_14344647 | 1.47 |
ENSDART00000188456
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr3_+_49521106 | 1.45 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr1_-_20273227 | 1.43 |
ENSDART00000146084
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr17_+_18117029 | 1.40 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr6_-_39270851 | 1.39 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr21_-_40782393 | 1.38 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr3_+_34919810 | 1.37 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr19_+_19729506 | 1.36 |
ENSDART00000171610
|
hoxa13a
|
homeobox A13a |
| chr23_+_13814978 | 1.35 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr12_+_15622621 | 1.35 |
ENSDART00000079784
|
plcd3b
|
phospholipase C, delta 3b |
| chr17_-_31058900 | 1.35 |
ENSDART00000134998
ENSDART00000104307 ENSDART00000172721 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr7_+_30970045 | 1.35 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr13_+_1089942 | 1.31 |
ENSDART00000054322
|
cnrip1b
|
cannabinoid receptor interacting protein 1b |
| chr8_+_53928472 | 1.30 |
ENSDART00000063242
|
MMP23B
|
matrix metallopeptidase 23B |
| chr11_-_15090118 | 1.29 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr1_-_26293203 | 1.26 |
ENSDART00000180140
|
cxxc4
|
CXXC finger 4 |
| chr17_-_51202339 | 1.25 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr3_+_18097700 | 1.25 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr7_-_50517023 | 1.24 |
ENSDART00000073910
|
adamtsl5
|
ADAMTS like 5 |
| chr14_-_29799993 | 1.24 |
ENSDART00000133775
ENSDART00000005568 |
pdlim3b
|
PDZ and LIM domain 3b |
| chr22_+_20427170 | 1.22 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr12_-_10275320 | 1.20 |
ENSDART00000170078
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr2_-_4797512 | 1.18 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr10_-_7988396 | 1.17 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr1_+_55080797 | 1.15 |
ENSDART00000077390
|
lgalsla
|
lectin, galactoside-binding-like a |
| chr22_-_910926 | 1.13 |
ENSDART00000180075
|
FP016205.1
|
|
| chr5_+_11407504 | 1.11 |
ENSDART00000186723
ENSDART00000018047 |
zgc:112294
|
zgc:112294 |
| chr25_+_19041329 | 1.10 |
ENSDART00000153467
|
lrtm2b
|
leucine-rich repeats and transmembrane domains 2b |
| chr7_-_35516251 | 1.10 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr15_-_18574716 | 1.08 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr1_-_45177373 | 1.06 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr8_+_23142946 | 1.06 |
ENSDART00000152933
|
si:ch211-196c10.13
|
si:ch211-196c10.13 |
| chr4_-_9173552 | 1.05 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr14_+_32839535 | 1.04 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr15_-_23467750 | 1.04 |
ENSDART00000148804
|
pdzd3a
|
PDZ domain containing 3a |
| chr25_+_19753890 | 1.01 |
ENSDART00000190268
ENSDART00000128852 |
celsr1b
|
cadherin EGF LAG seven-pass G-type receptor 1b |
| chr8_-_22965214 | 1.01 |
ENSDART00000148178
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr21_+_10739846 | 1.00 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr23_-_28294763 | 0.99 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr17_+_16564921 | 0.99 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr14_-_8080416 | 0.99 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr25_-_18913336 | 0.99 |
ENSDART00000171010
|
parietopsin
|
parietopsin |
| chr11_-_4235811 | 0.98 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr12_+_11080776 | 0.98 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr18_+_49969568 | 0.97 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr15_+_42599501 | 0.96 |
ENSDART00000177646
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr2_-_1569250 | 0.96 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr10_-_21054059 | 0.95 |
ENSDART00000139733
|
pcdh1a
|
protocadherin 1a |
| chr7_+_20471315 | 0.93 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr20_-_9963713 | 0.92 |
ENSDART00000104234
|
gjd2b
|
gap junction protein delta 2b |
| chr14_-_30587814 | 0.92 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr6_+_36795225 | 0.91 |
ENSDART00000171504
|
si:ch73-29l19.1
|
si:ch73-29l19.1 |
| chr15_+_37559570 | 0.90 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr9_-_18568927 | 0.89 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr12_+_39685485 | 0.89 |
ENSDART00000163403
|
LO017650.1
|
|
| chr7_+_57866292 | 0.88 |
ENSDART00000138757
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr8_+_14381272 | 0.87 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr21_+_32338897 | 0.84 |
ENSDART00000110137
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr17_-_20979077 | 0.82 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr1_-_39909985 | 0.81 |
ENSDART00000181673
|
stox2a
|
storkhead box 2a |
| chr3_-_28665291 | 0.81 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr9_-_8454060 | 0.80 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr13_-_47800342 | 0.79 |
ENSDART00000121817
|
fbln7
|
fibulin 7 |
| chr5_-_14390445 | 0.78 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr7_-_18656069 | 0.77 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr22_+_16446052 | 0.77 |
ENSDART00000142454
|
si:dkey-121a11.3
|
si:dkey-121a11.3 |
| chr14_+_909769 | 0.76 |
ENSDART00000021346
ENSDART00000172777 |
arl3l2
|
ADP-ribosylation factor-like 3, like 2 |
| chr23_-_24394719 | 0.74 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr5_+_43072951 | 0.73 |
ENSDART00000193905
ENSDART00000026020 |
wdr54
|
WD repeat domain 54 |
| chr15_-_15469079 | 0.71 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr7_-_49594995 | 0.71 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr1_-_17650223 | 0.70 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr6_+_32415132 | 0.69 |
ENSDART00000155790
|
kank4
|
KN motif and ankyrin repeat domains 4 |
| chr9_+_24095677 | 0.69 |
ENSDART00000150443
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr9_-_19161982 | 0.68 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr11_-_27702778 | 0.67 |
ENSDART00000045942
ENSDART00000125352 |
phf2
|
PHD finger protein 2 |
| chr16_+_24684107 | 0.65 |
ENSDART00000183920
|
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr15_+_29116063 | 0.65 |
ENSDART00000016112
ENSDART00000153609 ENSDART00000155630 |
capns1b
|
calpain, small subunit 1 b |
| chr10_-_36738619 | 0.64 |
ENSDART00000093000
ENSDART00000157179 |
PLEKHB1
|
si:ch211-176g13.7 |
| chr14_+_5861435 | 0.63 |
ENSDART00000041279
ENSDART00000147341 |
tubb4b
|
tubulin, beta 4B class IVb |
| chr17_+_25289431 | 0.63 |
ENSDART00000161002
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr23_-_35195908 | 0.62 |
ENSDART00000122429
|
klf15
|
Kruppel-like factor 15 |
| chr7_-_12821277 | 0.62 |
ENSDART00000091584
|
zgc:158785
|
zgc:158785 |
| chr3_+_34220194 | 0.62 |
ENSDART00000145859
|
slc25a23b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23b |
| chr5_+_34981584 | 0.61 |
ENSDART00000134795
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr4_-_22363709 | 0.61 |
ENSDART00000037670
|
orc5
|
origin recognition complex, subunit 5 |
| chr10_-_27199135 | 0.61 |
ENSDART00000189511
ENSDART00000180314 |
auts2a
|
autism susceptibility candidate 2a |
| chr24_-_1303935 | 0.61 |
ENSDART00000159212
ENSDART00000159267 ENSDART00000164904 |
nrp1a
|
neuropilin 1a |
| chr10_+_29260096 | 0.61 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr20_-_46541834 | 0.60 |
ENSDART00000060685
ENSDART00000181720 |
tmed10
|
transmembrane p24 trafficking protein 10 |
| chr11_-_29563437 | 0.60 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr6_+_9867426 | 0.59 |
ENSDART00000151749
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr23_-_27152866 | 0.59 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr2_+_30878864 | 0.58 |
ENSDART00000009326
|
oprk1
|
opioid receptor, kappa 1 |
| chr11_-_3860199 | 0.58 |
ENSDART00000082420
|
gata2a
|
GATA binding protein 2a |
| chr8_+_27743550 | 0.56 |
ENSDART00000046004
|
wnt2bb
|
wingless-type MMTV integration site family, member 2Bb |
| chr1_-_58009216 | 0.56 |
ENSDART00000143829
|
nxnl1
|
nucleoredoxin like 1 |
| chr14_-_25034142 | 0.54 |
ENSDART00000000392
|
pde6a
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr16_+_19536614 | 0.54 |
ENSDART00000112894
ENSDART00000079201 ENSDART00000139357 |
sp8b
|
sp8 transcription factor b |
| chr23_+_7505943 | 0.53 |
ENSDART00000135787
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr15_-_15468326 | 0.52 |
ENSDART00000161192
|
rab34a
|
RAB34, member RAS oncogene family a |
| chr12_+_25640480 | 0.52 |
ENSDART00000105608
|
prkcea
|
protein kinase C, epsilon a |
| chr7_-_52842605 | 0.51 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr1_+_33383644 | 0.50 |
ENSDART00000187194
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr20_+_35998073 | 0.50 |
ENSDART00000140457
|
opn5
|
opsin 5 |
| chr25_-_16818195 | 0.49 |
ENSDART00000185215
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr11_+_2506516 | 0.49 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr11_-_16215143 | 0.48 |
ENSDART00000027014
|
rab7
|
RAB7, member RAS oncogene family |
| chr2_-_48171441 | 0.48 |
ENSDART00000123040
|
pfkpb
|
phosphofructokinase, platelet b |
| chr15_-_24883956 | 0.48 |
ENSDART00000113199
|
aipl1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr17_+_36627099 | 0.46 |
ENSDART00000154104
|
impg1b
|
interphotoreceptor matrix proteoglycan 1b |
| chr20_-_1141722 | 0.46 |
ENSDART00000152675
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr25_-_7925269 | 0.45 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr3_+_15296824 | 0.44 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr17_+_16565185 | 0.44 |
ENSDART00000136874
|
foxn3
|
forkhead box N3 |
| chr18_+_5172848 | 0.43 |
ENSDART00000190642
|
CABZ01080599.1
|
|
| chr1_-_16507812 | 0.41 |
ENSDART00000169081
|
mtmr7b
|
myotubularin related protein 7b |
| chr25_-_7925019 | 0.40 |
ENSDART00000183309
|
glcea
|
glucuronic acid epimerase a |
| chr15_-_38154616 | 0.40 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr11_+_39672874 | 0.38 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr7_+_38395197 | 0.37 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr24_+_24923166 | 0.37 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr11_-_3860534 | 0.37 |
ENSDART00000082425
|
gata2a
|
GATA binding protein 2a |
| chr2_-_16217344 | 0.36 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr9_+_33357011 | 0.36 |
ENSDART00000088569
|
nyx
|
nyctalopin |
| chr7_+_41303572 | 0.36 |
ENSDART00000012692
|
rgs9bp
|
regulator of G protein signaling 9 binding protein |
| chr2_-_41861040 | 0.36 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr4_-_18436899 | 0.35 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr13_+_12528043 | 0.35 |
ENSDART00000057761
|
rrh
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr25_-_35996141 | 0.34 |
ENSDART00000149074
|
sall1b
|
spalt-like transcription factor 1b |
| chr18_+_7594012 | 0.34 |
ENSDART00000062150
|
zgc:77752
|
zgc:77752 |
| chr20_+_6543674 | 0.34 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr14_-_2400668 | 0.33 |
ENSDART00000172717
ENSDART00000182882 |
si:ch73-233f7.1
|
si:ch73-233f7.1 |
| chrM_+_15308 | 0.33 |
ENSDART00000093625
|
mt-cyb
|
cytochrome b, mitochondrial |
| chr2_+_28995776 | 0.33 |
ENSDART00000138733
|
cdh12a
|
cadherin 12, type 2a (N-cadherin 2) |
| chr2_-_48171112 | 0.33 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr24_-_31194847 | 0.32 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr3_-_61593274 | 0.31 |
ENSDART00000154132
ENSDART00000055071 |
nptx2a
|
neuronal pentraxin 2a |
| chr2_+_29795833 | 0.31 |
ENSDART00000056750
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr14_-_2933185 | 0.30 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr12_+_41991635 | 0.28 |
ENSDART00000186161
ENSDART00000192510 |
TCERG1L
|
transcription elongation regulator 1 like |
| chr16_-_17188294 | 0.27 |
ENSDART00000165883
|
opn9
|
opsin 9 |
| chr7_+_19903924 | 0.26 |
ENSDART00000159112
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr18_+_9323211 | 0.25 |
ENSDART00000166114
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr9_+_11034967 | 0.25 |
ENSDART00000152081
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr7_-_35515931 | 0.25 |
ENSDART00000193324
|
irx6a
|
iroquois homeobox 6a |
| chr13_-_36184701 | 0.23 |
ENSDART00000146522
|
map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr11_+_30296332 | 0.23 |
ENSDART00000192843
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr7_+_19904136 | 0.23 |
ENSDART00000173452
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr14_-_32089117 | 0.21 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr9_+_13714379 | 0.19 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr1_-_53987766 | 0.18 |
ENSDART00000167720
|
RHOU (1 of many)
|
si:ch211-133l11.10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of otx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0031446 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 2.3 | 6.9 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 1.1 | 3.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.7 | 2.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.5 | 4.6 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.5 | 1.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.5 | 8.6 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.4 | 2.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 3.1 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 1.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.3 | 0.9 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.3 | 3.4 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.3 | 1.2 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.3 | 1.5 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.2 | 0.7 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 1.9 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 2.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.2 | 0.9 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 0.5 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.2 | 2.1 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.2 | 1.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 1.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 1.3 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 0.6 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.8 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 1.9 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.3 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.6 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 1.0 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.5 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.5 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 1.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 3.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.8 | GO:0071632 | optomotor response(GO:0071632) |
| 0.1 | 2.7 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 1.0 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 1.3 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 1.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.8 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.3 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.4 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.6 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 2.1 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 1.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.4 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 1.6 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 1.0 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.0 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.5 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.8 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 2.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 5.1 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 1.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.0 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 1.2 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 3.5 | GO:0060041 | retina development in camera-type eye(GO:0060041) |
| 0.0 | 0.1 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 1.7 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.3 | 4.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 3.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 1.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 5.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.6 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 3.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 4.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 3.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 8.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.6 | 10.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.6 | 1.9 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.5 | 3.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.3 | 3.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 1.0 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.3 | 3.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 1.9 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.9 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.2 | 1.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 1.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.2 | 0.9 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 1.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.2 | 1.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 1.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 3.3 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 5.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.6 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 6.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 3.2 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 4.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.9 | GO:0015293 | symporter activity(GO:0015293) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 4.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 2.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.6 | 10.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 4.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 4.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |