Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
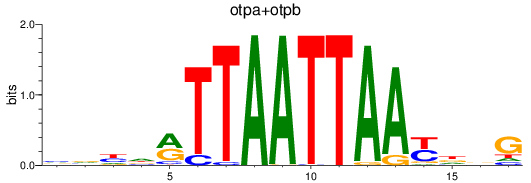
Results for otpa+otpb
Z-value: 0.52
Transcription factors associated with otpa+otpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
otpa
|
ENSDARG00000014201 | orthopedia homeobox a |
|
otpb
|
ENSDARG00000058379 | orthopedia homeobox b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otpa | dr11_v1_chr21_+_7582036_7582059 | 0.38 | 3.1e-01 | Click! |
| otpb | dr11_v1_chr5_-_51619742_51619742 | 0.31 | 4.2e-01 | Click! |
Activity profile of otpa+otpb motif
Sorted Z-values of otpa+otpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_35830625 | 1.57 |
ENSDART00000180028
|
CU459056.1
|
|
| chr17_+_15433518 | 1.09 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433671 | 1.03 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr21_+_25236297 | 0.96 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr25_+_21833287 | 0.87 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr4_+_9669717 | 0.66 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr2_-_30668580 | 0.63 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr1_-_669717 | 0.61 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr3_-_15999501 | 0.60 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr5_+_67390115 | 0.58 |
ENSDART00000193255
|
ebf2
|
early B cell factor 2 |
| chr3_-_19368435 | 0.56 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr14_+_46313396 | 0.55 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr7_-_30174882 | 0.53 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr1_-_31534089 | 0.52 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr17_+_41992054 | 0.48 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr9_+_37152564 | 0.48 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr6_+_25261297 | 0.47 |
ENSDART00000162824
ENSDART00000163490 ENSDART00000157790 ENSDART00000160978 ENSDART00000161545 ENSDART00000159978 |
kyat3
|
kynurenine aminotransferase 3 |
| chr15_-_21014270 | 0.46 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr4_+_57881965 | 0.46 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr9_-_22099536 | 0.46 |
ENSDART00000101923
|
CR391987.1
|
|
| chr1_+_21731382 | 0.46 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr6_+_25257728 | 0.46 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr20_-_45812144 | 0.45 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr7_+_38811800 | 0.44 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr14_-_413273 | 0.40 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr5_-_38451082 | 0.40 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr20_+_11731039 | 0.38 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr20_+_98179 | 0.37 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr12_+_20587179 | 0.37 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr19_+_43359075 | 0.35 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr11_+_34760628 | 0.35 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr1_-_25966068 | 0.33 |
ENSDART00000137869
ENSDART00000134192 |
synpo2b
|
synaptopodin 2b |
| chr8_-_33381430 | 0.33 |
ENSDART00000190019
|
lmx1bb
|
LIM homeobox transcription factor 1, beta b |
| chr19_+_43297546 | 0.33 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr15_+_5360407 | 0.33 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr23_+_11285662 | 0.33 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr15_-_22074315 | 0.32 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr23_-_16485190 | 0.31 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr1_-_5455498 | 0.31 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr12_-_31794908 | 0.31 |
ENSDART00000105583
ENSDART00000153449 |
nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr21_+_22840246 | 0.30 |
ENSDART00000151621
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr11_-_1509773 | 0.30 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr4_-_9891874 | 0.29 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr9_-_22834860 | 0.27 |
ENSDART00000146486
|
neb
|
nebulin |
| chr16_+_46111849 | 0.27 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr8_-_33381913 | 0.25 |
ENSDART00000076420
|
lmx1bb
|
LIM homeobox transcription factor 1, beta b |
| chr2_+_2223837 | 0.25 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr7_+_20512419 | 0.25 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr6_+_52350443 | 0.25 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr14_+_1170968 | 0.24 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr16_+_54209504 | 0.23 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr1_-_25966411 | 0.23 |
ENSDART00000193375
|
synpo2b
|
synaptopodin 2b |
| chr18_-_48547564 | 0.23 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr5_-_45877387 | 0.22 |
ENSDART00000183714
ENSDART00000041503 |
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr17_-_21418340 | 0.22 |
ENSDART00000007021
|
atp6v1ba
|
ATPase, H+ transporting, lysosomal, V1 subunit B, member a |
| chr3_-_16039619 | 0.22 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr7_+_72441515 | 0.22 |
ENSDART00000167827
|
CU459012.1
|
|
| chr3_+_6469754 | 0.22 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr14_-_451555 | 0.21 |
ENSDART00000190906
|
FAT4
|
FAT atypical cadherin 4 |
| chr1_+_45663727 | 0.21 |
ENSDART00000038574
ENSDART00000141144 ENSDART00000149565 |
trappc5
|
trafficking protein particle complex 5 |
| chr6_-_11768198 | 0.21 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr9_-_12811936 | 0.19 |
ENSDART00000188490
|
myo10l3
|
myosin X-like 3 |
| chr15_-_40267485 | 0.19 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr23_-_16484383 | 0.19 |
ENSDART00000187839
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr16_+_28932038 | 0.19 |
ENSDART00000149480
|
npr1b
|
natriuretic peptide receptor 1b |
| chr13_-_29420885 | 0.19 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr25_-_28674739 | 0.18 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr11_+_44617021 | 0.18 |
ENSDART00000158887
|
rbm34
|
RNA binding motif protein 34 |
| chr17_+_49281597 | 0.17 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr24_-_7995960 | 0.17 |
ENSDART00000186594
|
bloc1s5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr15_-_14552101 | 0.17 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr17_+_24821627 | 0.16 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr10_+_42423318 | 0.16 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr17_+_50524573 | 0.16 |
ENSDART00000187974
|
CR382385.1
|
|
| chr4_+_13449775 | 0.16 |
ENSDART00000172552
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr12_+_31713239 | 0.16 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr15_-_39955785 | 0.15 |
ENSDART00000154556
|
msh5
|
mutS homolog 5 |
| chr18_-_17485419 | 0.15 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr22_-_36530902 | 0.15 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr1_+_51039558 | 0.15 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr22_-_19552796 | 0.15 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr9_-_42418470 | 0.15 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr3_+_3681116 | 0.14 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr23_+_11347313 | 0.14 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr3_+_26342768 | 0.13 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr14_-_858985 | 0.13 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr5_+_71802014 | 0.13 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr9_+_34127005 | 0.13 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr5_-_12272689 | 0.13 |
ENSDART00000099810
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr23_-_1660708 | 0.13 |
ENSDART00000175138
|
CU693481.1
|
|
| chr5_-_45876908 | 0.12 |
ENSDART00000188229
|
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr4_+_54618332 | 0.12 |
ENSDART00000171824
|
si:ch211-227e10.2
|
si:ch211-227e10.2 |
| chr11_+_40812590 | 0.12 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr7_-_71829649 | 0.12 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr9_-_25055722 | 0.12 |
ENSDART00000137131
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr6_+_39905021 | 0.11 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr1_+_11882186 | 0.11 |
ENSDART00000193419
|
snx8b
|
sorting nexin 8b |
| chr2_+_20605925 | 0.11 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr9_-_32177117 | 0.10 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr10_-_11353469 | 0.10 |
ENSDART00000191847
|
lin54
|
lin-54 DREAM MuvB core complex component |
| chr25_+_13621563 | 0.09 |
ENSDART00000159428
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr9_+_28598577 | 0.09 |
ENSDART00000142623
ENSDART00000135947 |
si:ch73-7i4.1
|
si:ch73-7i4.1 |
| chr3_+_13848226 | 0.09 |
ENSDART00000184342
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr2_+_29842878 | 0.09 |
ENSDART00000131376
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr2_-_37140423 | 0.09 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr8_-_13046089 | 0.09 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr14_-_1355544 | 0.08 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr22_-_14247276 | 0.08 |
ENSDART00000033332
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr6_+_24420523 | 0.08 |
ENSDART00000185461
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr17_+_31914877 | 0.07 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr25_-_27722614 | 0.07 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr10_+_11261576 | 0.07 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr4_-_1801519 | 0.07 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr4_+_59845617 | 0.06 |
ENSDART00000167626
ENSDART00000123157 |
si:dkey-196n19.2
|
si:dkey-196n19.2 |
| chr21_-_35534401 | 0.06 |
ENSDART00000112308
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr7_-_52388734 | 0.06 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr11_-_40742424 | 0.06 |
ENSDART00000173399
ENSDART00000021369 |
tas1r3
|
taste receptor, type 1, member 3 |
| chr20_+_34717403 | 0.06 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr5_+_2815021 | 0.06 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr4_-_67980261 | 0.06 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr8_-_44926077 | 0.06 |
ENSDART00000084417
|
timm17b
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr1_+_51721851 | 0.06 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr6_-_40029423 | 0.05 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr24_+_28953089 | 0.05 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr13_-_12602920 | 0.05 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr1_-_19502322 | 0.04 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr17_-_2386569 | 0.04 |
ENSDART00000121614
|
PLCB2
|
phospholipase C beta 2 |
| chr11_-_2838699 | 0.04 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr13_-_52089003 | 0.04 |
ENSDART00000187600
|
tmem254
|
transmembrane protein 254 |
| chr6_+_14980761 | 0.04 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr24_-_35282568 | 0.04 |
ENSDART00000167406
ENSDART00000088609 |
sntg1
|
syntrophin, gamma 1 |
| chr18_+_7456597 | 0.04 |
ENSDART00000142270
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr9_+_23895711 | 0.04 |
ENSDART00000034686
|
cops8
|
COP9 signalosome subunit 8 |
| chr17_+_16564921 | 0.03 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr10_+_26747755 | 0.03 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr11_-_10456387 | 0.03 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr6_-_33878665 | 0.03 |
ENSDART00000129916
|
tmem69
|
transmembrane protein 69 |
| chr2_+_37140448 | 0.03 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr5_+_60590796 | 0.03 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr22_-_9183944 | 0.03 |
ENSDART00000188599
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr4_+_19700308 | 0.03 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr14_+_989733 | 0.02 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr15_+_37965953 | 0.02 |
ENSDART00000154192
|
si:dkey-238d18.10
|
si:dkey-238d18.10 |
| chr18_-_5527050 | 0.02 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr18_+_17725410 | 0.02 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr22_+_508290 | 0.02 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr18_+_18439332 | 0.02 |
ENSDART00000043843
|
siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr16_+_40024883 | 0.00 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr18_+_33272861 | 0.00 |
ENSDART00000131796
|
v2ra20
|
vomeronasal 2 receptor, a20 |
| chr1_+_9086942 | 0.00 |
ENSDART00000055023
|
cacng3a
|
calcium channel, voltage-dependent, gamma subunit 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of otpa+otpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.3 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.9 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.2 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.5 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.6 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.0 | 0.6 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.1 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:0010387 | protein deneddylation(GO:0000338) COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.9 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |