Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for osr1
Z-value: 1.89
Transcription factors associated with osr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
osr1
|
ENSDARG00000014091 | odd-skipped related transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| osr1 | dr11_v1_chr13_+_32144370_32144370 | 0.68 | 4.5e-02 | Click! |
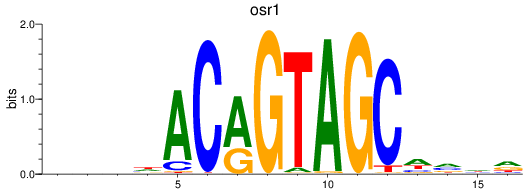
Activity profile of osr1 motif
Sorted Z-values of osr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_29954709 | 4.83 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chr3_-_55139127 | 4.46 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr7_+_29955368 | 4.36 |
ENSDART00000173686
|
tpma
|
alpha-tropomyosin |
| chr14_-_26536504 | 3.88 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr9_-_45601103 | 3.67 |
ENSDART00000180465
|
agr1
|
anterior gradient 1 |
| chr1_+_17593392 | 3.41 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
| chr9_-_22069364 | 3.11 |
ENSDART00000101938
|
crygm2b
|
crystallin, gamma M2b |
| chr3_-_61181018 | 2.96 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr18_-_45736 | 2.89 |
ENSDART00000148373
ENSDART00000148950 |
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr18_-_46010 | 2.85 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr14_+_51098036 | 2.84 |
ENSDART00000184118
|
CABZ01078594.1
|
|
| chr9_-_22023061 | 2.42 |
ENSDART00000101952
|
crygm2c
|
crystallin, gamma M2c |
| chr2_+_56463167 | 2.36 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr1_-_58064738 | 2.36 |
ENSDART00000073778
|
caspb
|
caspase b |
| chr7_+_44713135 | 2.21 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr1_-_40911332 | 2.07 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr7_-_27033080 | 2.03 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr3_-_37758487 | 2.03 |
ENSDART00000150938
|
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr3_-_29899172 | 2.01 |
ENSDART00000140518
ENSDART00000020311 |
rpl27
|
ribosomal protein L27 |
| chr15_-_39848822 | 1.94 |
ENSDART00000155230
|
si:dkey-263j23.1
|
si:dkey-263j23.1 |
| chr23_-_31403668 | 1.90 |
ENSDART00000147498
|
CR759830.1
|
|
| chr13_-_992458 | 1.88 |
ENSDART00000114655
|
BFSP1
|
beaded filament structural protein 1 |
| chr19_+_31576849 | 1.81 |
ENSDART00000134107
ENSDART00000088401 |
acot13
|
acyl-CoA thioesterase 13 |
| chr17_+_31185276 | 1.80 |
ENSDART00000062887
|
disp2
|
dispatched homolog 2 (Drosophila) |
| chr18_-_46258612 | 1.73 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr18_+_30847237 | 1.71 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr11_+_14199802 | 1.70 |
ENSDART00000102520
ENSDART00000133172 |
palm1a
|
paralemmin 1a |
| chr10_+_19554604 | 1.68 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr13_-_1408775 | 1.66 |
ENSDART00000049684
|
bag2
|
BCL2 associated athanogene 2 |
| chr18_-_977075 | 1.64 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr5_-_24201437 | 1.60 |
ENSDART00000114113
|
sox19a
|
SRY (sex determining region Y)-box 19a |
| chr11_+_2198831 | 1.57 |
ENSDART00000160515
|
hoxc6b
|
homeobox C6b |
| chr7_+_26534131 | 1.55 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr16_-_26537103 | 1.55 |
ENSDART00000134908
ENSDART00000008152 |
sgk2b
|
serum/glucocorticoid regulated kinase 2b |
| chr22_-_5006801 | 1.53 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr10_-_25860102 | 1.51 |
ENSDART00000080789
|
trpc4a
|
transient receptor potential cation channel, subfamily C, member 4a |
| chr14_+_4151379 | 1.48 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr16_-_46660680 | 1.47 |
ENSDART00000159209
ENSDART00000191929 |
tmem176l.4
|
transmembrane protein 176l.4 |
| chr5_-_56513825 | 1.47 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr22_-_28871787 | 1.46 |
ENSDART00000104828
|
gtpbp2b
|
GTP binding protein 2b |
| chr22_-_11626014 | 1.45 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr1_-_22861348 | 1.44 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr13_+_28417297 | 1.42 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr20_-_29787192 | 1.40 |
ENSDART00000125348
ENSDART00000048759 |
id2b
|
inhibitor of DNA binding 2b |
| chr5_-_36949476 | 1.39 |
ENSDART00000047269
|
h3f3c
|
H3 histone, family 3C |
| chr14_-_40390757 | 1.39 |
ENSDART00000149443
|
pcdh19
|
protocadherin 19 |
| chr10_-_13239367 | 1.37 |
ENSDART00000001253
|
si:busm1-57f23.1
|
si:busm1-57f23.1 |
| chr7_+_22767678 | 1.35 |
ENSDART00000137203
|
ponzr6
|
plac8 onzin related protein 6 |
| chr14_-_24391424 | 1.34 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr20_-_9436521 | 1.34 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr2_-_11512819 | 1.32 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr2_-_9646857 | 1.32 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr23_-_46040618 | 1.31 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr12_-_38548299 | 1.28 |
ENSDART00000153374
|
si:dkey-1f1.3
|
si:dkey-1f1.3 |
| chr24_+_10413484 | 1.28 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr1_-_21723329 | 1.28 |
ENSDART00000137138
|
si:ch211-134c9.2
|
si:ch211-134c9.2 |
| chr13_-_11536951 | 1.27 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr1_-_21344478 | 1.26 |
ENSDART00000077805
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr21_+_33454147 | 1.24 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr21_-_15674802 | 1.23 |
ENSDART00000136666
|
mmp11b
|
matrix metallopeptidase 11b |
| chr2_-_37896965 | 1.22 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr20_-_5291012 | 1.21 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr10_+_38512270 | 1.19 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr2_-_51096647 | 1.18 |
ENSDART00000167172
|
THEM6
|
si:ch73-52e5.2 |
| chr12_+_31713239 | 1.18 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr19_-_35361556 | 1.17 |
ENSDART00000012167
|
ndufs5
|
NADH dehydrogenase (ubiquinone) Fe-S protein 5 |
| chr5_-_48307804 | 1.17 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr18_+_46382484 | 1.16 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr1_+_23783349 | 1.16 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr20_-_29418620 | 1.14 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr1_-_59130695 | 1.12 |
ENSDART00000152560
|
FP015850.1
|
|
| chr9_-_33121535 | 1.11 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr2_-_30668580 | 1.09 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr5_+_69808763 | 1.09 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr3_-_30123113 | 1.08 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr11_+_21053488 | 1.08 |
ENSDART00000189860
|
zgc:113307
|
zgc:113307 |
| chr11_-_33960318 | 1.07 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr22_-_21314821 | 1.07 |
ENSDART00000105546
ENSDART00000135388 |
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr4_+_47257854 | 1.06 |
ENSDART00000173868
|
crestin
|
crestin |
| chr13_-_32635859 | 1.05 |
ENSDART00000146249
ENSDART00000145395 ENSDART00000148040 ENSDART00000100650 |
matn3b
|
matrilin 3b |
| chr10_+_21559605 | 1.05 |
ENSDART00000123648
ENSDART00000108584 |
pcdh1a3
pcdh1a3
|
protocadherin 1 alpha 3 protocadherin 1 alpha 3 |
| chr4_-_6567355 | 1.04 |
ENSDART00000134820
ENSDART00000142087 |
foxp2
|
forkhead box P2 |
| chr19_-_25519310 | 1.02 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr18_+_21408794 | 1.01 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr3_-_53722182 | 1.01 |
ENSDART00000032788
|
rdh8a
|
retinol dehydrogenase 8a |
| chr16_-_28876479 | 0.99 |
ENSDART00000047862
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr8_+_4798158 | 0.99 |
ENSDART00000031650
|
hsp70l
|
heat shock cognate 70-kd protein, like |
| chr8_-_2506327 | 0.98 |
ENSDART00000101125
ENSDART00000125124 |
rpl6
|
ribosomal protein L6 |
| chr2_-_20574193 | 0.98 |
ENSDART00000190448
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr13_+_30696286 | 0.98 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr16_+_35344031 | 0.97 |
ENSDART00000167140
|
si:dkey-34d22.1
|
si:dkey-34d22.1 |
| chr12_-_2993095 | 0.97 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr7_+_33314925 | 0.97 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr25_+_34581494 | 0.96 |
ENSDART00000167690
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr7_+_31871830 | 0.96 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr4_-_1824836 | 0.96 |
ENSDART00000111858
|
mrpl42
|
mitochondrial ribosomal protein L42 |
| chr20_+_1412193 | 0.95 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr13_-_45523026 | 0.95 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr9_-_18568927 | 0.95 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr4_+_3287819 | 0.94 |
ENSDART00000168633
|
CABZ01085700.1
|
|
| chr14_-_17072736 | 0.93 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr21_+_4702773 | 0.93 |
ENSDART00000147404
|
si:dkey-102g19.3
|
si:dkey-102g19.3 |
| chr14_+_29769336 | 0.92 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr18_-_49116382 | 0.91 |
ENSDART00000174157
|
BX663503.3
|
|
| chr22_+_661711 | 0.88 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr14_-_13048355 | 0.88 |
ENSDART00000166434
|
si:dkey-35h6.1
|
si:dkey-35h6.1 |
| chr19_-_9712530 | 0.87 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr25_-_31433512 | 0.87 |
ENSDART00000067028
|
zgc:172145
|
zgc:172145 |
| chr18_+_30507839 | 0.86 |
ENSDART00000026866
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr19_+_14921000 | 0.85 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr9_-_12424231 | 0.84 |
ENSDART00000188952
|
zgc:162707
|
zgc:162707 |
| chr11_-_36350876 | 0.83 |
ENSDART00000146495
ENSDART00000020655 |
psma5
|
proteasome subunit alpha 5 |
| chr20_+_30490682 | 0.83 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr12_-_45669050 | 0.82 |
ENSDART00000124455
|
fbxl15
|
F-box and leucine-rich repeat protein 15 |
| chr4_+_8569199 | 0.82 |
ENSDART00000165181
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr4_-_23908802 | 0.81 |
ENSDART00000138873
|
celf2
|
cugbp, Elav-like family member 2 |
| chr8_-_50981175 | 0.81 |
ENSDART00000004065
|
zgc:91909
|
zgc:91909 |
| chr14_+_35901249 | 0.78 |
ENSDART00000105604
|
zgc:77938
|
zgc:77938 |
| chr4_-_67969695 | 0.78 |
ENSDART00000190016
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr23_-_9925568 | 0.77 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr6_-_19035749 | 0.75 |
ENSDART00000187714
|
sept9b
|
septin 9b |
| chr19_-_5769553 | 0.74 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr19_-_25519612 | 0.74 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr7_+_48805534 | 0.74 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr14_+_49135264 | 0.73 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr9_-_5263947 | 0.72 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr23_-_30785382 | 0.72 |
ENSDART00000136156
ENSDART00000131285 |
myt1a
|
myelin transcription factor 1a |
| chr9_-_306515 | 0.71 |
ENSDART00000166059
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr5_-_5831037 | 0.71 |
ENSDART00000112856
|
zmp:0000000846
|
zmp:0000000846 |
| chr2_+_20539402 | 0.70 |
ENSDART00000129585
|
si:ch73-14h1.2
|
si:ch73-14h1.2 |
| chr2_-_9748039 | 0.69 |
ENSDART00000134870
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr20_+_16881883 | 0.68 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr7_-_4036875 | 0.67 |
ENSDART00000165021
|
ndrg2
|
NDRG family member 2 |
| chr15_-_12011202 | 0.67 |
ENSDART00000160427
ENSDART00000168715 |
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr21_+_45627775 | 0.67 |
ENSDART00000185466
|
irf1b
|
interferon regulatory factor 1b |
| chr12_-_45876387 | 0.67 |
ENSDART00000043210
ENSDART00000149044 |
pax2b
|
paired box 2b |
| chr7_-_59564011 | 0.66 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr6_-_48087152 | 0.65 |
ENSDART00000180614
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr3_+_15805917 | 0.65 |
ENSDART00000055834
|
phospho1
|
phosphatase, orphan 1 |
| chr5_-_55981288 | 0.65 |
ENSDART00000146616
|
si:dkey-189h5.6
|
si:dkey-189h5.6 |
| chr3_-_26115558 | 0.64 |
ENSDART00000078671
ENSDART00000124762 |
hsp70.1
|
heat shock cognate 70-kd protein, tandem duplicate 1 |
| chr3_-_38918697 | 0.62 |
ENSDART00000145630
|
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr2_+_22531185 | 0.62 |
ENSDART00000171959
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr13_-_27038407 | 0.62 |
ENSDART00000146712
|
ccdc85a
|
coiled-coil domain containing 85A |
| chr21_-_40083432 | 0.60 |
ENSDART00000141160
ENSDART00000191195 |
slc13a5a
|
info solute carrier family 13 (sodium-dependent citrate transporter), member 5a |
| chr1_+_52563298 | 0.60 |
ENSDART00000142465
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr8_+_49778756 | 0.58 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr4_-_12323228 | 0.58 |
ENSDART00000081089
|
il17ra1a
|
interleukin 17 receptor A1a |
| chr24_-_9960290 | 0.58 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr2_-_37896646 | 0.57 |
ENSDART00000075931
|
hbl1
|
hexose-binding lectin 1 |
| chr1_-_8140763 | 0.56 |
ENSDART00000091508
|
FAM83G
|
si:dkeyp-9d4.4 |
| chr1_+_54650048 | 0.55 |
ENSDART00000141207
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr13_+_18663600 | 0.55 |
ENSDART00000141647
|
selenou1a
|
selenoprotein U 1a |
| chr17_-_22021213 | 0.54 |
ENSDART00000078843
|
zmp:0000001102
|
zmp:0000001102 |
| chr15_-_35960250 | 0.54 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr21_-_26490186 | 0.53 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr10_+_2587234 | 0.52 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr9_-_21098413 | 0.52 |
ENSDART00000018570
|
wars2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr14_+_5383060 | 0.52 |
ENSDART00000187825
|
lbx2
|
ladybird homeobox 2 |
| chr13_+_50151407 | 0.52 |
ENSDART00000031858
|
gpr137ba
|
G protein-coupled receptor 137Ba |
| chr24_+_1294176 | 0.52 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr3_+_19460991 | 0.50 |
ENSDART00000169124
|
pde6ga
|
phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog a |
| chr14_+_17137023 | 0.50 |
ENSDART00000080712
|
slc43a3b
|
solute carrier family 43, member 3b |
| chr13_+_40686133 | 0.49 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr2_-_4868002 | 0.49 |
ENSDART00000189835
|
tfr1a
|
transferrin receptor 1a |
| chr12_-_7824291 | 0.49 |
ENSDART00000148673
ENSDART00000149453 |
ank3b
|
ankyrin 3b |
| chr14_-_17305849 | 0.48 |
ENSDART00000160971
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr2_+_5446882 | 0.48 |
ENSDART00000083260
|
dusp28
|
dual specificity phosphatase 28 |
| chr11_+_31285127 | 0.48 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr16_+_20871021 | 0.47 |
ENSDART00000006429
|
hibadhb
|
3-hydroxyisobutyrate dehydrogenase b |
| chr20_+_10727022 | 0.47 |
ENSDART00000104185
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr14_+_22680485 | 0.46 |
ENSDART00000167829
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr7_+_48805725 | 0.46 |
ENSDART00000166543
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr12_+_48815988 | 0.46 |
ENSDART00000149089
|
anxa11b
|
annexin A11b |
| chr21_-_25295087 | 0.46 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr6_-_8407406 | 0.46 |
ENSDART00000151355
|
ptger1a
|
prostaglandin E receptor 1a (subtype EP1) |
| chr14_-_17306261 | 0.45 |
ENSDART00000191747
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr7_+_20656942 | 0.45 |
ENSDART00000100898
|
tnfsf12
|
TNF superfamily member 12 |
| chr13_+_35746440 | 0.45 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr15_-_576135 | 0.45 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr17_-_6198823 | 0.44 |
ENSDART00000028407
ENSDART00000193636 |
ptk2ba
|
protein tyrosine kinase 2 beta, a |
| chr9_-_4856767 | 0.44 |
ENSDART00000016814
ENSDART00000138015 |
fmnl2a
|
formin-like 2a |
| chr17_-_45104750 | 0.43 |
ENSDART00000075520
|
aldh6a1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr1_+_9886991 | 0.43 |
ENSDART00000135702
|
rgs11
|
regulator of G protein signaling 11 |
| chr15_+_3219134 | 0.42 |
ENSDART00000113532
|
LO017656.1
|
|
| chr23_+_7379728 | 0.42 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr3_-_32958505 | 0.42 |
ENSDART00000147374
ENSDART00000136919 |
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr3_-_16285861 | 0.41 |
ENSDART00000140103
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr21_-_40834413 | 0.41 |
ENSDART00000148513
|
limk1b
|
LIM domain kinase 1b |
| chr2_+_3986083 | 0.41 |
ENSDART00000188979
|
mkxb
|
mohawk homeobox b |
| chr6_+_49082796 | 0.40 |
ENSDART00000182446
|
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr10_+_3875716 | 0.39 |
ENSDART00000189268
ENSDART00000180624 |
TTC28
|
tetratricopeptide repeat domain 28 |
| chr11_-_36279602 | 0.39 |
ENSDART00000122531
ENSDART00000103064 ENSDART00000125616 |
nfya
|
nuclear transcription factor Y, alpha |
| chr25_-_13659249 | 0.39 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr8_+_30709685 | 0.38 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr6_-_41229787 | 0.38 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr20_+_36010080 | 0.38 |
ENSDART00000021456
|
opn5
|
opsin 5 |
| chr19_-_26823647 | 0.37 |
ENSDART00000002464
|
neu1
|
neuraminidase 1 |
| chr6_-_40884453 | 0.37 |
ENSDART00000017968
ENSDART00000154100 |
sirt4
|
sirtuin 4 |
| chr20_+_53522059 | 0.37 |
ENSDART00000147570
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
Network of associatons between targets according to the STRING database.
First level regulatory network of osr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.5 | 1.5 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.4 | 1.2 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.3 | 1.3 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 0.9 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.2 | 0.7 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.2 | 1.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 0.9 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 4.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 2.0 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.2 | 0.6 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 1.0 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 1.0 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 0.5 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 2.1 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 0.9 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.6 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 1.0 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 1.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.9 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.5 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 1.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 1.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 1.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.8 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.1 | 1.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.4 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 1.6 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.1 | 1.0 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.1 | 0.4 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 1.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 9.4 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.1 | 0.5 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 1.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.4 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0048913 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.1 | 1.0 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.2 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.2 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 1.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.7 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.1 | 0.2 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.6 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 1.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 5.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:1901380 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.8 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 1.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 3.9 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.7 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 1.6 | GO:0035195 | gene silencing by miRNA(GO:0035195) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 1.4 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 1.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 1.3 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.2 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 0.6 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 2.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 2.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.6 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.3 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 2.1 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.1 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 3.7 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.6 | 2.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.4 | 4.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.5 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 1.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 9.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 1.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 1.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 5.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.7 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 4.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.3 | 1.4 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.3 | 1.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.3 | 0.9 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.3 | 1.0 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 1.1 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 1.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.2 | 2.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 0.6 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.2 | 1.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 5.7 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 1.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 0.8 | GO:0051903 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.1 | 5.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.6 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.5 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 1.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.5 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.1 | 1.8 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.9 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 5.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 9.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.3 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 1.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.5 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 3.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 4.9 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 3.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 7.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 4.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |