Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
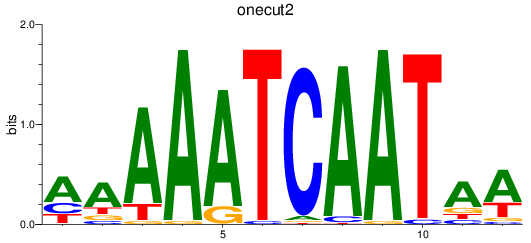
Results for onecut2
Z-value: 0.76
Transcription factors associated with onecut2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
onecut2
|
ENSDARG00000090387 | one cut homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| onecut2 | dr11_v1_chr21_-_1742159_1742159 | 0.26 | 4.9e-01 | Click! |
Activity profile of onecut2 motif
Sorted Z-values of onecut2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_17526735 | 0.72 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr11_-_11518469 | 0.65 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr6_+_52804267 | 0.63 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr11_+_43114108 | 0.61 |
ENSDART00000013642
ENSDART00000190266 |
foxg1b
|
forkhead box G1b |
| chr7_+_15266093 | 0.61 |
ENSDART00000124676
|
sv2ba
|
synaptic vesicle glycoprotein 2Ba |
| chr1_-_21409877 | 0.60 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr8_-_23081511 | 0.58 |
ENSDART00000142015
ENSDART00000135764 ENSDART00000147021 |
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr3_-_28120092 | 0.58 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr8_-_14484599 | 0.55 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr6_+_3828560 | 0.54 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr4_+_13810811 | 0.53 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr11_+_37201483 | 0.52 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr6_-_10828880 | 0.51 |
ENSDART00000131458
ENSDART00000020261 |
chrna1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr1_+_16144615 | 0.49 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr25_-_9805269 | 0.48 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr15_-_26552393 | 0.47 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr23_+_28770225 | 0.47 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr1_-_23157583 | 0.47 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr5_-_29643930 | 0.45 |
ENSDART00000161250
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr10_-_7785930 | 0.44 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr7_+_20535869 | 0.41 |
ENSDART00000078181
|
zgc:158423
|
zgc:158423 |
| chr23_-_17470146 | 0.41 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr8_+_37168570 | 0.40 |
ENSDART00000098636
|
csf1b
|
colony stimulating factor 1b (macrophage) |
| chr16_-_16701718 | 0.38 |
ENSDART00000143550
|
si:dkey-8k3.2
|
si:dkey-8k3.2 |
| chr4_-_10599062 | 0.38 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr5_+_61301525 | 0.38 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr21_-_20342096 | 0.37 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr19_-_32641725 | 0.37 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr21_-_20341836 | 0.37 |
ENSDART00000176689
|
rbp4l
|
retinol binding protein 4, like |
| chr19_-_41472228 | 0.36 |
ENSDART00000113388
|
dlx5a
|
distal-less homeobox 5a |
| chr10_-_33156789 | 0.36 |
ENSDART00000192268
ENSDART00000182065 ENSDART00000081170 |
cux1a
|
cut-like homeobox 1a |
| chr13_+_28417297 | 0.34 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr12_+_26467847 | 0.34 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr15_-_26552652 | 0.34 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr10_+_10801719 | 0.33 |
ENSDART00000193648
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr24_+_25692802 | 0.33 |
ENSDART00000190493
|
camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr23_-_15216654 | 0.33 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr10_-_28761454 | 0.32 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr3_-_46811611 | 0.32 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr18_+_50276653 | 0.32 |
ENSDART00000192120
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr4_-_5291256 | 0.31 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr18_+_50276337 | 0.30 |
ENSDART00000140352
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr3_-_28665291 | 0.28 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr16_+_22587661 | 0.28 |
ENSDART00000129612
ENSDART00000142241 |
she
|
Src homology 2 domain containing E |
| chr2_-_32505091 | 0.28 |
ENSDART00000141884
ENSDART00000056639 |
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr3_-_22228602 | 0.28 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr14_+_35024521 | 0.28 |
ENSDART00000158634
ENSDART00000170631 |
ebf3a
|
early B cell factor 3a |
| chr18_+_642889 | 0.28 |
ENSDART00000189007
|
CABZ01078320.1
|
|
| chr25_+_7229046 | 0.27 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr1_+_37391716 | 0.27 |
ENSDART00000191986
|
sparcl1
|
SPARC-like 1 |
| chr21_-_27881752 | 0.27 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr2_+_7818368 | 0.26 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr19_+_4916233 | 0.26 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr3_+_23687909 | 0.25 |
ENSDART00000046638
|
hoxb8a
|
homeobox B8a |
| chr4_-_2868112 | 0.25 |
ENSDART00000133843
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr10_-_15405564 | 0.25 |
ENSDART00000020665
|
sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr20_-_19422496 | 0.25 |
ENSDART00000143658
|
si:ch211-278j3.3
|
si:ch211-278j3.3 |
| chr14_-_2348917 | 0.24 |
ENSDART00000159004
|
si:ch73-233f7.8
|
si:ch73-233f7.8 |
| chr4_+_26628822 | 0.24 |
ENSDART00000191030
ENSDART00000186113 ENSDART00000186764 ENSDART00000165158 |
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr4_-_8903240 | 0.23 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr21_-_28901095 | 0.22 |
ENSDART00000180820
|
cxxc5a
|
CXXC finger protein 5a |
| chr5_+_4366431 | 0.22 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr5_-_63286077 | 0.22 |
ENSDART00000131274
|
tbcelb
|
tubulin folding cofactor E-like b |
| chr24_+_2962988 | 0.22 |
ENSDART00000164449
|
eci2
|
enoyl-CoA delta isomerase 2 |
| chr17_-_45247332 | 0.21 |
ENSDART00000016815
|
ttbk2a
|
tau tubulin kinase 2a |
| chr16_-_26255877 | 0.21 |
ENSDART00000146214
|
erfl1
|
Ets2 repressor factor like 1 |
| chr2_+_45300512 | 0.21 |
ENSDART00000144704
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr4_+_7827261 | 0.21 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr6_-_7052595 | 0.21 |
ENSDART00000081761
ENSDART00000181351 |
bin1b
|
bridging integrator 1b |
| chr2_+_5563077 | 0.21 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr2_-_39759059 | 0.20 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr18_+_48428126 | 0.20 |
ENSDART00000137978
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr18_-_37007294 | 0.19 |
ENSDART00000088309
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr5_+_56268436 | 0.19 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr8_-_4010887 | 0.19 |
ENSDART00000163678
|
mtmr3
|
myotubularin related protein 3 |
| chr18_+_36769758 | 0.19 |
ENSDART00000180375
ENSDART00000136463 ENSDART00000133487 ENSDART00000130206 |
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr20_-_40755614 | 0.19 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr17_+_8175998 | 0.18 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr12_-_43428542 | 0.17 |
ENSDART00000192266
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr5_+_27267186 | 0.17 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr14_+_21699414 | 0.17 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr18_+_50890749 | 0.17 |
ENSDART00000174109
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr18_+_15644559 | 0.17 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr2_+_22042745 | 0.16 |
ENSDART00000132039
|
tox
|
thymocyte selection-associated high mobility group box |
| chr15_-_5467477 | 0.16 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr4_-_8902406 | 0.16 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr18_-_37007061 | 0.15 |
ENSDART00000136432
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr5_-_65158203 | 0.15 |
ENSDART00000171656
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr19_+_42886413 | 0.15 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr9_+_44431174 | 0.15 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr20_-_6812688 | 0.15 |
ENSDART00000170934
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr8_-_32385989 | 0.14 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr5_+_19309877 | 0.14 |
ENSDART00000190338
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr15_+_27384798 | 0.13 |
ENSDART00000164887
|
tbx4
|
T-box 4 |
| chr4_-_13255700 | 0.13 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr25_+_19955598 | 0.13 |
ENSDART00000091547
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr9_+_44430974 | 0.13 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr7_-_71434298 | 0.13 |
ENSDART00000180507
|
lgi2a
|
leucine-rich repeat LGI family, member 2a |
| chr16_+_32729223 | 0.12 |
ENSDART00000125663
|
si:dkey-165n16.1
|
si:dkey-165n16.1 |
| chr23_-_19434199 | 0.12 |
ENSDART00000132543
|
klhdc8b
|
kelch domain containing 8B |
| chr4_+_16787488 | 0.12 |
ENSDART00000143006
|
golt1ba
|
golgi transport 1Ba |
| chr21_+_19070921 | 0.12 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr6_-_7052408 | 0.12 |
ENSDART00000150033
ENSDART00000149232 |
bin1b
|
bridging integrator 1b |
| chr8_-_45279411 | 0.12 |
ENSDART00000175207
|
adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr16_+_46492994 | 0.12 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr20_+_52546186 | 0.12 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr2_-_6262441 | 0.12 |
ENSDART00000092190
|
arl14
|
ADP-ribosylation factor-like 14 |
| chr8_+_40477264 | 0.11 |
ENSDART00000085559
|
gck
|
glucokinase (hexokinase 4) |
| chr15_-_34785594 | 0.11 |
ENSDART00000154256
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr7_+_49877148 | 0.11 |
ENSDART00000174182
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr8_-_2616326 | 0.11 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr9_+_44430705 | 0.11 |
ENSDART00000190696
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr3_-_8399774 | 0.11 |
ENSDART00000190238
ENSDART00000113140 ENSDART00000132949 |
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr18_-_17030870 | 0.11 |
ENSDART00000079817
|
has3
|
hyaluronan synthase 3 |
| chr13_-_24877577 | 0.10 |
ENSDART00000182705
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr15_+_41901970 | 0.10 |
ENSDART00000152724
|
si:ch211-191a16.5
|
si:ch211-191a16.5 |
| chr18_-_21806613 | 0.10 |
ENSDART00000145721
|
nrn1la
|
neuritin 1-like a |
| chr3_+_39853788 | 0.10 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr2_-_31735142 | 0.10 |
ENSDART00000130903
|
ralyl
|
RALY RNA binding protein like |
| chr25_-_21085661 | 0.09 |
ENSDART00000099355
|
prr5a
|
proline rich 5a (renal) |
| chr5_+_42136359 | 0.09 |
ENSDART00000083731
|
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr16_-_2650341 | 0.09 |
ENSDART00000128169
ENSDART00000155432 ENSDART00000103722 |
lyplal1
|
lysophospholipase-like 1 |
| chr13_-_31008275 | 0.09 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr17_-_8169774 | 0.09 |
ENSDART00000091828
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr17_+_41081977 | 0.09 |
ENSDART00000137091
ENSDART00000143714 |
babam2
|
BRISC and BRCA1 A complex member 2 |
| chr19_-_18855513 | 0.09 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr4_-_9214288 | 0.09 |
ENSDART00000067309
|
avpr1ab
|
arginine vasopressin receptor 1Ab |
| chr6_-_2627488 | 0.09 |
ENSDART00000044089
ENSDART00000158333 ENSDART00000155109 |
hyi
|
hydroxypyruvate isomerase |
| chr21_-_28439596 | 0.09 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr19_-_18855717 | 0.08 |
ENSDART00000158192
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr11_-_426525 | 0.08 |
ENSDART00000157054
|
znf831
|
zinc finger protein 831 |
| chr14_+_21699129 | 0.08 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr22_-_7129631 | 0.08 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr5_-_28102698 | 0.08 |
ENSDART00000138140
|
si:ch211-48m9.1
|
si:ch211-48m9.1 |
| chr15_+_17446796 | 0.08 |
ENSDART00000157189
|
snx19b
|
sorting nexin 19b |
| chr1_-_54191626 | 0.07 |
ENSDART00000062941
|
nthl1
|
nth-like DNA glycosylase 1 |
| chr18_-_46380471 | 0.07 |
ENSDART00000144444
ENSDART00000086772 |
slc19a3b
|
solute carrier family 19 (thiamine transporter), member 3b |
| chr6_-_16456093 | 0.07 |
ENSDART00000083305
ENSDART00000181640 |
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr7_+_42206847 | 0.07 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr7_+_25036188 | 0.07 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr17_+_6957042 | 0.06 |
ENSDART00000103839
|
zgc:172341
|
zgc:172341 |
| chr8_+_9866351 | 0.06 |
ENSDART00000133985
|
kcnd1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr15_+_21947117 | 0.05 |
ENSDART00000156234
|
ttc12
|
tetratricopeptide repeat domain 12 |
| chr24_-_36680261 | 0.05 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr8_+_35664152 | 0.05 |
ENSDART00000144520
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr21_-_7687544 | 0.05 |
ENSDART00000134519
|
pde8b
|
phosphodiesterase 8B |
| chr21_+_20229926 | 0.04 |
ENSDART00000025860
|
si:dkey-247m21.3
|
si:dkey-247m21.3 |
| chr17_+_40989973 | 0.04 |
ENSDART00000160049
|
mrpl33
|
mitochondrial ribosomal protein L33 |
| chr20_-_40754794 | 0.04 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr13_+_36144341 | 0.04 |
ENSDART00000182930
ENSDART00000187327 |
TTC9
|
si:ch211-259k16.3 |
| chr21_-_22812417 | 0.04 |
ENSDART00000079151
ENSDART00000112175 |
trpc6a
|
transient receptor potential cation channel, subfamily C, member 6a |
| chr13_-_30713236 | 0.04 |
ENSDART00000112372
ENSDART00000142221 |
tmem72
|
transmembrane protein 72 |
| chr21_+_17005737 | 0.04 |
ENSDART00000101246
|
vps29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr21_+_16145980 | 0.03 |
ENSDART00000135119
|
qrfpr4
|
pyroglutamylated RFamide peptide receptor 4 |
| chr3_-_60142530 | 0.02 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr12_-_25217217 | 0.02 |
ENSDART00000152931
|
kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr16_+_3067134 | 0.02 |
ENSDART00000012048
|
cyb5r4
|
cytochrome b5 reductase 4 |
| chr11_-_22605981 | 0.02 |
ENSDART00000186923
|
myog
|
myogenin |
| chr9_-_7287128 | 0.02 |
ENSDART00000176281
ENSDART00000065803 |
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr21_-_41369370 | 0.01 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr1_-_28861226 | 0.00 |
ENSDART00000075502
|
timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr9_-_7287375 | 0.00 |
ENSDART00000128352
|
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr17_+_21760032 | 0.00 |
ENSDART00000190425
|
ikzf5
|
IKAROS family zinc finger 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of onecut2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 0.7 | GO:0046551 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.2 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.1 | 0.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.5 | GO:0086009 | membrane repolarization(GO:0086009) |
| 0.1 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.6 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.2 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.2 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.5 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.6 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.0 | 0.4 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:1900120 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.4 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.4 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.0 | 0.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.2 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.3 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.6 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 0.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 0.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.5 | GO:0042166 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.1 | 0.7 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |