Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
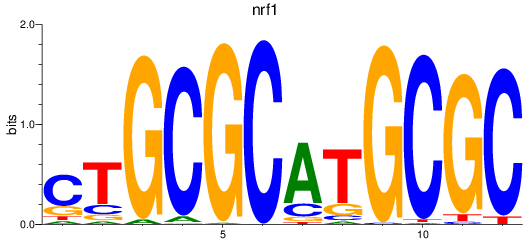
Results for nrf1
Z-value: 5.08
Transcription factors associated with nrf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nrf1
|
ENSDARG00000000018 | nuclear respiratory factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nrf1 | dr11_v1_chr4_-_15103646_15103696 | 0.71 | 3.1e-02 | Click! |
Activity profile of nrf1 motif
Sorted Z-values of nrf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_22320738 | 6.92 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr25_+_22319940 | 6.11 |
ENSDART00000154065
ENSDART00000153492 ENSDART00000024866 ENSDART00000154376 |
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr13_-_2336584 | 5.36 |
ENSDART00000113692
|
tceb3l
|
transcription elongation factor B (SIII), polypeptide 3, like |
| chr5_+_62319217 | 5.32 |
ENSDART00000050879
|
pho
|
phoenix |
| chr15_+_44201056 | 5.17 |
ENSDART00000162433
ENSDART00000148336 |
CU655961.4
|
|
| chr12_-_30359498 | 5.04 |
ENSDART00000152981
ENSDART00000189988 |
tdrd1
|
tudor domain containing 1 |
| chr24_-_42072886 | 4.94 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr12_-_30359031 | 4.91 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr15_-_45538773 | 4.88 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr11_-_6452444 | 4.78 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr2_+_29997043 | 4.67 |
ENSDART00000139566
ENSDART00000151848 |
rbm33b
|
RNA binding motif protein 33b |
| chr15_-_28587147 | 4.57 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr2_+_29996650 | 4.54 |
ENSDART00000138050
ENSDART00000141026 |
rbm33b
|
RNA binding motif protein 33b |
| chr12_+_47081783 | 4.52 |
ENSDART00000158568
|
mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr7_+_57089354 | 4.45 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr20_-_182841 | 4.26 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr23_+_45845159 | 4.24 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr14_+_14806851 | 4.15 |
ENSDART00000169235
|
fhdc2
|
FH2 domain containing 2 |
| chr12_-_9468618 | 4.14 |
ENSDART00000152737
ENSDART00000091519 |
pgap3
|
post-GPI attachment to proteins 3 |
| chr13_+_20524921 | 4.11 |
ENSDART00000081385
|
ccdc172
|
coiled-coil domain containing 172 |
| chr25_+_6266009 | 4.10 |
ENSDART00000148995
|
slc25a44a
|
solute carrier family 25, member 44 a |
| chr17_+_51940768 | 4.04 |
ENSDART00000053422
|
ttll5
|
tubulin tyrosine ligase-like family, member 5 |
| chr15_+_44184367 | 3.98 |
ENSDART00000162918
ENSDART00000110060 |
zgc:165514
|
zgc:165514 |
| chr7_-_37895771 | 3.87 |
ENSDART00000084282
|
papd5
|
PAP associated domain containing 5 |
| chr8_+_7875110 | 3.80 |
ENSDART00000167423
ENSDART00000160267 ENSDART00000180490 |
mbd1a
|
methyl-CpG binding domain protein 1a |
| chr13_-_24826607 | 3.79 |
ENSDART00000087786
ENSDART00000186951 |
slka
|
STE20-like kinase a |
| chr22_-_38274188 | 3.76 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr5_-_18961694 | 3.75 |
ENSDART00000142531
ENSDART00000090521 |
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr25_-_37121335 | 3.71 |
ENSDART00000017805
|
nfat5a
|
nuclear factor of activated T cells 5a |
| chr1_+_51615672 | 3.66 |
ENSDART00000165117
|
zgc:165656
|
zgc:165656 |
| chr22_+_21549419 | 3.63 |
ENSDART00000139411
|
plpp2b
|
phospholipid phosphatase 2b |
| chr19_-_18127808 | 3.62 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr8_+_50150834 | 3.61 |
ENSDART00000056074
|
entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr19_-_18127629 | 3.58 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr8_-_22326744 | 3.53 |
ENSDART00000137645
|
cep104
|
centrosomal protein 104 |
| chr8_+_23861461 | 3.52 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr5_+_19933356 | 3.47 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr7_+_46020508 | 3.45 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr18_-_7097403 | 3.43 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr1_+_51191049 | 3.43 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr11_+_27134116 | 3.42 |
ENSDART00000129736
|
hdac11
|
histone deacetylase 11 |
| chr17_-_868004 | 3.41 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr14_-_46198373 | 3.40 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr10_+_29855213 | 3.38 |
ENSDART00000099992
ENSDART00000127723 ENSDART00000125475 |
jhy
|
junctional cadherin complex regulator |
| chr13_-_15793585 | 3.38 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2 associated athanogene 5 |
| chr15_+_44250335 | 3.37 |
ENSDART00000186162
ENSDART00000193503 ENSDART00000180275 |
zgc:162962
|
zgc:162962 |
| chr14_-_763744 | 3.37 |
ENSDART00000165856
|
trim35-27
|
tripartite motif containing 35-27 |
| chr21_+_3928947 | 3.34 |
ENSDART00000149777
|
setx
|
senataxin |
| chr25_+_2776511 | 3.31 |
ENSDART00000115280
|
neo1b
|
neogenin 1b |
| chr5_+_29652513 | 3.29 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr25_-_20666754 | 3.29 |
ENSDART00000158418
|
csk
|
C-terminal Src kinase |
| chr19_+_28291062 | 3.29 |
ENSDART00000163382
|
lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr7_+_57088920 | 3.27 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr19_-_4785734 | 3.26 |
ENSDART00000113088
|
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr19_+_28291376 | 3.21 |
ENSDART00000139433
ENSDART00000103855 |
lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr23_+_45845423 | 3.20 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr12_-_33558727 | 3.19 |
ENSDART00000086087
|
mbtd1
|
mbt domain containing 1 |
| chr2_-_17492486 | 3.19 |
ENSDART00000189464
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr11_+_5565082 | 3.19 |
ENSDART00000112590
ENSDART00000183207 |
si:ch73-40i7.5
|
si:ch73-40i7.5 |
| chr19_+_4856351 | 3.18 |
ENSDART00000093402
|
cdk12
|
cyclin-dependent kinase 12 |
| chr23_-_44723102 | 3.12 |
ENSDART00000129138
|
mogat3a
|
monoacylglycerol O-acyltransferase 3a |
| chr11_+_27133560 | 3.12 |
ENSDART00000158411
|
hdac11
|
histone deacetylase 11 |
| chr10_-_40939303 | 3.10 |
ENSDART00000134295
|
bmp1b
|
bone morphogenetic protein 1b |
| chr12_-_33558879 | 3.10 |
ENSDART00000161167
|
mbtd1
|
mbt domain containing 1 |
| chr19_+_30450125 | 3.09 |
ENSDART00000073704
|
si:ch211-215a10.4
|
si:ch211-215a10.4 |
| chr22_-_4989803 | 3.05 |
ENSDART00000181359
ENSDART00000125265 ENSDART00000028634 ENSDART00000183294 |
cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr18_+_14645568 | 3.05 |
ENSDART00000138995
ENSDART00000147351 |
vps9d1
|
VPS9 domain containing 1 |
| chr14_+_14806692 | 3.05 |
ENSDART00000193050
|
fhdc2
|
FH2 domain containing 2 |
| chr2_+_30182431 | 3.04 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr14_-_899170 | 3.04 |
ENSDART00000165211
ENSDART00000031992 |
rgs14a
|
regulator of G protein signaling 14a |
| chr6_-_9676108 | 3.03 |
ENSDART00000169915
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr3_+_48473346 | 3.01 |
ENSDART00000166294
|
metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr9_+_37366973 | 2.98 |
ENSDART00000016370
|
dirc2
|
disrupted in renal carcinoma 2 |
| chr19_+_24891747 | 2.95 |
ENSDART00000132209
ENSDART00000193610 |
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr23_-_26227805 | 2.94 |
ENSDART00000158082
|
BX927204.1
|
|
| chr23_+_40410644 | 2.94 |
ENSDART00000056328
|
elovl4b
|
ELOVL fatty acid elongase 4b |
| chr23_-_26228077 | 2.93 |
ENSDART00000162423
|
BX927204.1
|
|
| chr25_+_8921425 | 2.93 |
ENSDART00000128591
|
accs
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| chr6_-_12900154 | 2.93 |
ENSDART00000080408
ENSDART00000150887 |
ical1
|
islet cell autoantigen 1-like |
| chr16_+_25285998 | 2.90 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr19_-_7043355 | 2.89 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr21_+_11521163 | 2.88 |
ENSDART00000139267
|
zgc:114104
|
zgc:114104 |
| chr21_-_18275226 | 2.88 |
ENSDART00000126672
ENSDART00000135239 |
brd3a
|
bromodomain containing 3a |
| chr6_-_7686594 | 2.86 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr21_+_38745094 | 2.85 |
ENSDART00000113316
|
heatr6
|
HEAT repeat containing 6 |
| chr11_-_30508843 | 2.84 |
ENSDART00000101667
ENSDART00000179930 |
map4k3a
|
mitogen-activated protein kinase kinase kinase kinase 3a |
| chr25_+_2776865 | 2.82 |
ENSDART00000156567
|
neo1b
|
neogenin 1b |
| chr11_+_31236001 | 2.82 |
ENSDART00000129393
|
trmt1
|
tRNA methyltransferase 1 |
| chr13_+_25364324 | 2.81 |
ENSDART00000187471
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr21_-_3613702 | 2.81 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr20_-_6131686 | 2.80 |
ENSDART00000145964
ENSDART00000086578 ENSDART00000164090 |
pum2
|
pumilio RNA-binding family member 2 |
| chr7_+_17953589 | 2.79 |
ENSDART00000174778
ENSDART00000113120 |
taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr19_-_42238003 | 2.78 |
ENSDART00000151022
|
si:ch211-191i18.4
|
si:ch211-191i18.4 |
| chr10_-_40939706 | 2.77 |
ENSDART00000059795
ENSDART00000190510 |
bmp1b
|
bone morphogenetic protein 1b |
| chr2_-_17492080 | 2.77 |
ENSDART00000024302
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr13_+_25364753 | 2.75 |
ENSDART00000027428
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr1_-_53918839 | 2.75 |
ENSDART00000032552
|
taf5l
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr4_-_12286067 | 2.74 |
ENSDART00000022646
|
cnot4b
|
CCR4-NOT transcription complex, subunit 4b |
| chr12_+_2804505 | 2.74 |
ENSDART00000152193
|
mms19
|
MMS19 homolog, cytosolic iron-sulfur assembly component |
| chr25_-_34740627 | 2.74 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr3_+_14571813 | 2.73 |
ENSDART00000146728
ENSDART00000171731 |
znf653
|
zinc finger protein 653 |
| chr13_-_35844961 | 2.73 |
ENSDART00000171371
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr16_-_32304764 | 2.72 |
ENSDART00000143859
ENSDART00000134381 |
mms22l
|
MMS22-like, DNA repair protein |
| chr25_-_35296165 | 2.72 |
ENSDART00000018107
|
fancf
|
Fanconi anemia, complementation group F |
| chr12_-_2800809 | 2.71 |
ENSDART00000152682
ENSDART00000083784 |
ubtd1b
|
ubiquitin domain containing 1b |
| chr5_+_431994 | 2.71 |
ENSDART00000181692
ENSDART00000170350 |
thap1
|
THAP domain containing, apoptosis associated protein 1 |
| chr25_-_35101673 | 2.70 |
ENSDART00000140864
|
zgc:162611
|
zgc:162611 |
| chr7_+_39738505 | 2.69 |
ENSDART00000004365
|
tada2b
|
transcriptional adaptor 2B |
| chr23_-_31810222 | 2.69 |
ENSDART00000134319
ENSDART00000139076 |
hbs1l
|
HBS1-like translational GTPase |
| chr25_-_37186894 | 2.68 |
ENSDART00000191647
ENSDART00000182095 |
tdrd12
|
tudor domain containing 12 |
| chr21_-_11327830 | 2.68 |
ENSDART00000122331
|
rtkn2b
|
rhotekin 2b |
| chr2_-_58075414 | 2.67 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr22_-_547748 | 2.67 |
ENSDART00000037455
ENSDART00000140101 |
ccnd3
|
cyclin D3 |
| chr23_-_27442544 | 2.67 |
ENSDART00000019521
|
dip2ba
|
disco-interacting protein 2 homolog Ba |
| chr9_+_25096500 | 2.66 |
ENSDART00000135074
ENSDART00000180436 ENSDART00000108629 |
lrch1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr17_+_33415319 | 2.65 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr14_+_46118834 | 2.65 |
ENSDART00000124417
ENSDART00000017785 |
naa15a
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit a |
| chr1_-_39983730 | 2.62 |
ENSDART00000160066
|
ing2
|
inhibitor of growth family, member 2 |
| chr1_-_50247 | 2.62 |
ENSDART00000168428
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr5_+_36666715 | 2.61 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr20_-_14114078 | 2.61 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr12_+_36428052 | 2.60 |
ENSDART00000131300
|
unk
|
unkempt family zinc finger |
| chr5_+_29652198 | 2.59 |
ENSDART00000184083
|
tsc1a
|
TSC complex subunit 1a |
| chr2_+_21356242 | 2.59 |
ENSDART00000145670
ENSDART00000146600 |
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr24_+_39129316 | 2.58 |
ENSDART00000155346
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr4_-_5691257 | 2.57 |
ENSDART00000110497
|
tmem63a
|
transmembrane protein 63A |
| chr21_+_6114709 | 2.56 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr23_+_43668756 | 2.56 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr20_+_38543542 | 2.55 |
ENSDART00000145254
|
gtf3c2
|
general transcription factor IIIC, polypeptide 2, beta |
| chr25_+_32496877 | 2.54 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr25_+_15938880 | 2.54 |
ENSDART00000089035
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr24_-_25166416 | 2.53 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr13_+_39277178 | 2.52 |
ENSDART00000113259
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr1_+_19094548 | 2.52 |
ENSDART00000114514
|
ptpn9b
|
protein tyrosine phosphatase, non-receptor type 9, b |
| chr13_+_49727333 | 2.52 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr21_-_4764120 | 2.51 |
ENSDART00000129355
ENSDART00000102643 |
camsap1a
|
calmodulin regulated spectrin-associated protein 1a |
| chr21_+_6114305 | 2.49 |
ENSDART00000141607
|
fpgs
|
folylpolyglutamate synthase |
| chr23_+_12160900 | 2.49 |
ENSDART00000136046
|
ppp1r3da
|
protein phosphatase 1, regulatory subunit 3Da |
| chr18_+_17600570 | 2.47 |
ENSDART00000175258
ENSDART00000151850 ENSDART00000151934 |
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr5_+_71924175 | 2.47 |
ENSDART00000115182
ENSDART00000170215 |
nup214
|
nucleoporin 214 |
| chr23_+_43770149 | 2.46 |
ENSDART00000024313
|
rnf150b
|
ring finger protein 150b |
| chr8_-_34427364 | 2.45 |
ENSDART00000112854
ENSDART00000161282 ENSDART00000113230 |
gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr8_-_51954562 | 2.44 |
ENSDART00000132527
ENSDART00000057315 |
cep78
|
centrosomal protein 78 |
| chr6_-_24392909 | 2.44 |
ENSDART00000171042
ENSDART00000168355 |
brdt
|
bromodomain, testis-specific |
| chr1_+_47165842 | 2.43 |
ENSDART00000053152
ENSDART00000167051 |
cbr1
|
carbonyl reductase 1 |
| chr15_-_16384184 | 2.43 |
ENSDART00000154504
|
fam222bb
|
family with sequence similarity 222, member Bb |
| chr21_-_11367271 | 2.43 |
ENSDART00000151000
ENSDART00000151465 |
zgc:162472
|
zgc:162472 |
| chr19_-_42238440 | 2.42 |
ENSDART00000132591
|
si:ch211-191i18.4
|
si:ch211-191i18.4 |
| chr13_-_48161568 | 2.42 |
ENSDART00000109469
ENSDART00000188052 ENSDART00000193446 ENSDART00000189509 ENSDART00000184810 |
golga4
|
golgin A4 |
| chr2_+_21356500 | 2.41 |
ENSDART00000171397
|
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr5_+_23913585 | 2.40 |
ENSDART00000015401
|
ercc6l
|
excision repair cross-complementation group 6-like |
| chr6_-_18228358 | 2.39 |
ENSDART00000167937
|
p4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr10_-_17550931 | 2.38 |
ENSDART00000145077
|
ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr4_+_2482046 | 2.38 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr19_-_47832853 | 2.38 |
ENSDART00000170988
|
ago4
|
argonaute RISC catalytic component 4 |
| chr2_-_57076687 | 2.37 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr13_-_9367647 | 2.37 |
ENSDART00000083362
ENSDART00000144146 |
si:dkey-33c12.4
|
si:dkey-33c12.4 |
| chr25_-_35101396 | 2.36 |
ENSDART00000138865
|
zgc:162611
|
zgc:162611 |
| chr15_+_38221038 | 2.36 |
ENSDART00000188149
|
stim1a
|
stromal interaction molecule 1a |
| chr10_-_3394256 | 2.35 |
ENSDART00000158315
ENSDART00000111241 |
naa25
|
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
| chr21_+_18274825 | 2.35 |
ENSDART00000144322
ENSDART00000147768 |
wdr5
|
WD repeat domain 5 |
| chr21_+_3897680 | 2.35 |
ENSDART00000170653
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr21_-_34032650 | 2.33 |
ENSDART00000138575
ENSDART00000047515 |
rnf145b
|
ring finger protein 145b |
| chr21_-_1644414 | 2.32 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr15_+_24549054 | 2.31 |
ENSDART00000155900
|
phf12b
|
PHD finger protein 12b |
| chr5_-_11809404 | 2.31 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr8_-_45939691 | 2.29 |
ENSDART00000040066
ENSDART00000132297 |
adam9
|
ADAM metallopeptidase domain 9 |
| chr18_+_3579829 | 2.29 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr20_-_53624645 | 2.28 |
ENSDART00000083427
ENSDART00000152920 |
slc25a29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr5_-_2676783 | 2.27 |
ENSDART00000159661
|
CABZ01072548.1
|
|
| chr25_-_20666328 | 2.27 |
ENSDART00000098076
|
csk
|
C-terminal Src kinase |
| chr23_+_10805188 | 2.27 |
ENSDART00000035693
|
ppp4r2a
|
protein phosphatase 4, regulatory subunit 2a |
| chr21_-_14878220 | 2.27 |
ENSDART00000131237
|
ulk1b
|
unc-51 like autophagy activating kinase 1 |
| chr24_-_24983047 | 2.26 |
ENSDART00000066631
|
slc51a
|
solute carrier family 51, alpha subunit |
| chr1_+_15258641 | 2.26 |
ENSDART00000033018
|
pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr3_-_40837218 | 2.26 |
ENSDART00000134026
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr11_-_20988238 | 2.26 |
ENSDART00000155238
|
taf4a
|
TAF4A RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr5_+_47862636 | 2.26 |
ENSDART00000139824
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr11_+_2763168 | 2.25 |
ENSDART00000042972
|
srpk1b
|
SRSF protein kinase 1b |
| chr12_+_22560067 | 2.25 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr19_-_7070691 | 2.23 |
ENSDART00000168755
|
tapbp.2
|
TAP binding protein (tapasin), tandem duplicate 2 |
| chr16_-_26528140 | 2.22 |
ENSDART00000134448
ENSDART00000147062 |
l3mbtl1b
|
l(3)mbt-like 1b (Drosophila) |
| chr22_+_8536838 | 2.21 |
ENSDART00000132998
|
si:ch73-27e22.7
|
si:ch73-27e22.7 |
| chr13_-_49144799 | 2.20 |
ENSDART00000030939
ENSDART00000148922 |
disc1
|
disrupted in schizophrenia 1 |
| chr6_-_9695294 | 2.18 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr20_+_1121458 | 2.16 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr10_+_37268854 | 2.16 |
ENSDART00000131897
|
nf1b
|
neurofibromin 1b |
| chr2_+_2818645 | 2.16 |
ENSDART00000163587
|
rock1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr2_+_30489846 | 2.15 |
ENSDART00000145732
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr5_-_64454459 | 2.15 |
ENSDART00000172321
ENSDART00000168030 |
brd3b
|
bromodomain containing 3b |
| chr17_+_33415542 | 2.15 |
ENSDART00000183169
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr9_+_34380299 | 2.15 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr9_+_45605410 | 2.14 |
ENSDART00000136444
ENSDART00000007189 ENSDART00000158713 ENSDART00000182671 |
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr19_+_20787179 | 2.14 |
ENSDART00000193204
|
adnp2b
|
ADNP homeobox 2b |
| chr13_-_34862452 | 2.14 |
ENSDART00000134573
ENSDART00000047552 |
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr15_-_30984557 | 2.13 |
ENSDART00000080328
|
nf1a
|
neurofibromin 1a |
| chr24_-_25166720 | 2.13 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr12_+_18906407 | 2.12 |
ENSDART00000105854
|
josd1
|
Josephin domain containing 1 |
| chr11_+_2649891 | 2.10 |
ENSDART00000093052
|
si:ch211-160o17.4
|
si:ch211-160o17.4 |
| chr25_-_752158 | 2.09 |
ENSDART00000130610
|
tmem117
|
transmembrane protein 117 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nrf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 13.0 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 1.5 | 4.5 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 1.4 | 7.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 1.3 | 5.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 1.2 | 3.6 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 1.1 | 5.6 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 1.1 | 5.6 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 1.0 | 9.9 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.9 | 2.8 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.9 | 5.5 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.9 | 2.7 | GO:2000402 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.8 | 2.5 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.8 | 5.4 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.7 | 5.0 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.7 | 3.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.7 | 2.0 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.6 | 5.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.6 | 4.3 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.6 | 5.4 | GO:2000758 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.6 | 1.8 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.6 | 3.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.6 | 2.3 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.6 | 2.2 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.6 | 1.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.6 | 2.2 | GO:0021755 | eurydendroid cell differentiation(GO:0021755) |
| 0.5 | 2.7 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.5 | 1.6 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.5 | 2.0 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.5 | 1.8 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.5 | 1.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.5 | 3.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.4 | 5.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.4 | 2.1 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.4 | 5.9 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.4 | 3.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.4 | 1.6 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.4 | 2.0 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.4 | 2.4 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.4 | 2.0 | GO:0061438 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.4 | 1.9 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.4 | 1.8 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.4 | 1.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 5.8 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.3 | 2.7 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.3 | 1.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 1.9 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 1.5 | GO:0010939 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.3 | 4.5 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.3 | 1.2 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.3 | 2.0 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.3 | 2.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 0.9 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.3 | 0.8 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 5.3 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.3 | 2.9 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.3 | 1.3 | GO:0045056 | transcytosis(GO:0045056) |
| 0.3 | 0.8 | GO:0051170 | nuclear import(GO:0051170) |
| 0.2 | 3.0 | GO:0042572 | retinoic acid biosynthetic process(GO:0002138) retinol metabolic process(GO:0042572) |
| 0.2 | 4.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.2 | 3.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 6.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 1.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 4.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 4.0 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.2 | 5.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.2 | 2.1 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.2 | 4.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.2 | 3.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.2 | 2.8 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.2 | 2.0 | GO:0070977 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.2 | 1.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 2.2 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.2 | 1.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 2.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.2 | 1.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 1.3 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.2 | 0.5 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 2.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.2 | 5.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 0.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 3.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 0.8 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.2 | 1.0 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.2 | 2.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 3.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.2 | 1.3 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 0.5 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.2 | 2.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 1.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 1.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 2.3 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 2.5 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 4.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.4 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.8 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 1.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 1.9 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.7 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 6.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 1.3 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.1 | 2.0 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.7 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.1 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.1 | 3.0 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 2.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.3 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 1.3 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 5.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 2.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 3.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 1.5 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 2.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.6 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.8 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 2.0 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 1.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 2.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.6 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.7 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.9 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 3.1 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 1.4 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 2.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.6 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 1.9 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 2.5 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 0.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 1.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.3 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.1 | 0.9 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.1 | 1.8 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 7.4 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 3.0 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.1 | 3.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 2.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 1.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 1.6 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 1.9 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 1.4 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 2.1 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.6 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.1 | 4.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 0.4 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.1 | 0.5 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 5.2 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.1 | 8.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 1.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.7 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.1 | 1.2 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 1.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 4.1 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.0 | 1.0 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 4.1 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.4 | GO:0006901 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 3.8 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 2.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.0 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 8.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.2 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 1.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 3.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.5 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 2.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.4 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 2.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 2.0 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 2.1 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.2 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.0 | 1.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.9 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 1.1 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 1.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.7 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 10.1 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 0.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 5.2 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.7 | GO:0038127 | ERBB signaling pathway(GO:0038127) |
| 0.0 | 0.1 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.7 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.4 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.8 | GO:0045089 | positive regulation of innate immune response(GO:0045089) |
| 0.0 | 0.7 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.0 | 0.4 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.6 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 1.0 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.9 | GO:0071546 | pi-body(GO:0071546) |
| 1.2 | 5.9 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.1 | 6.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 1.1 | 1.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 1.0 | 3.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.9 | 5.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.9 | 2.7 | GO:0097361 | CIA complex(GO:0097361) |
| 0.8 | 5.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.8 | 5.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.8 | 2.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.7 | 4.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.6 | 3.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.6 | 2.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.6 | 1.7 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.5 | 3.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.5 | 2.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.5 | 5.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.5 | 2.4 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.4 | 1.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.4 | 1.2 | GO:0008352 | katanin complex(GO:0008352) |
| 0.4 | 7.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 4.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 3.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.4 | 1.8 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.3 | 1.4 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 2.4 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.3 | 2.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 0.9 | GO:0072380 | TRC complex(GO:0072380) |
| 0.3 | 0.8 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.3 | 2.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.3 | 1.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 2.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 2.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 5.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 2.7 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.2 | 4.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 2.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 3.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 3.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 2.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 1.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 3.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 1.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 1.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 1.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 1.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 3.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 7.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.4 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.5 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 2.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 2.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 6.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 2.0 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 3.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 4.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 14.6 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 3.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 9.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 3.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.0 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 3.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 3.6 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.1 | 1.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 4.2 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.3 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 1.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 6.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 11.9 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 2.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 5.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.1 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.9 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 3.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 1.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 2.1 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 7.7 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.5 | GO:0005657 | replication fork(GO:0005657) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 1.4 | 5.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.3 | 6.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 1.2 | 9.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 1.1 | 3.3 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.9 | 2.6 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.8 | 2.4 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.7 | 2.8 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.6 | 3.1 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.6 | 2.3 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.6 | 4.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.6 | 2.2 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.6 | 5.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.5 | 3.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.5 | 2.0 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.5 | 6.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.5 | 3.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.4 | 1.8 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.4 | 1.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 5.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 2.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 5.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 3.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.4 | 4.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 1.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 10.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.3 | 4.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 3.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 5.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.3 | 1.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 3.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 3.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.3 | 10.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.3 | 2.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 3.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 1.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 4.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.3 | 0.8 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.3 | 2.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.3 | 2.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.3 | 5.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.3 | 0.8 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.3 | 5.1 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.3 | 2.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.2 | 2.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 1.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 1.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 3.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 1.8 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.2 | 4.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 3.6 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 1.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 1.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 1.7 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.2 | 2.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 2.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 2.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 4.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.6 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 1.9 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.7 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 3.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 3.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.9 | GO:0005123 | death receptor binding(GO:0005123) receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 5.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 3.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 2.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.9 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 1.9 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 0.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.5 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 1.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 7.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 2.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 2.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 1.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 13.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 2.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 2.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 4.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.2 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.1 | 5.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 3.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 1.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 10.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.2 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.1 | 6.9 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 0.9 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 4.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 11.7 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 12.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 2.9 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 1.4 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 3.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.2 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 1.3 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 3.6 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 9.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.5 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.0 | 3.4 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 5.2 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 13.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 2.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 4.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 6.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.5 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 2.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 5.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 2.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 5.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 5.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 2.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 3.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 0.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 1.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 13.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 1.1 | 5.6 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.8 | 6.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.6 | 5.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.4 | 3.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 4.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.2 | 2.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 4.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 3.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 2.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 6.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 7.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 4.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 3.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 1.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 3.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 2.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 3.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.5 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.8 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.1 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |