Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
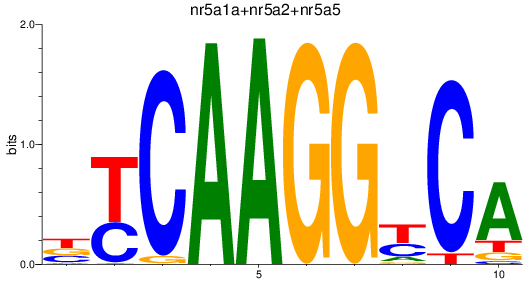
Results for nr5a1a+nr5a2+nr5a5
Z-value: 1.71
Transcription factors associated with nr5a1a+nr5a2+nr5a5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr5a5
|
ENSDARG00000039116 | nuclear receptor subfamily 5, group A, member 5 |
|
nr5a2
|
ENSDARG00000100940 | nuclear receptor subfamily 5, group A, member 2 |
|
nr5a1a
|
ENSDARG00000103176 | nuclear receptor subfamily 5, group A, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr5a1a | dr11_v1_chr8_-_52859301_52859301 | 0.87 | 2.1e-03 | Click! |
| nr5a2 | dr11_v1_chr22_-_22719440_22719440 | 0.75 | 1.9e-02 | Click! |
| nr5a5 | dr11_v1_chr3_-_53465223_53465249 | 0.59 | 9.6e-02 | Click! |
Activity profile of nr5a1a+nr5a2+nr5a5 motif
Sorted Z-values of nr5a1a+nr5a2+nr5a5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_1970071 | 4.75 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr19_-_30404096 | 3.80 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr15_-_23645810 | 3.73 |
ENSDART00000168845
|
ckmb
|
creatine kinase, muscle b |
| chr19_-_30403922 | 3.30 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr6_-_15653494 | 2.50 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr7_+_25913225 | 2.44 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr20_+_10539293 | 2.37 |
ENSDART00000182265
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr17_-_12385308 | 2.24 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr22_-_2886937 | 2.15 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr6_+_37894914 | 2.15 |
ENSDART00000148817
|
oca2
|
oculocutaneous albinism II |
| chr21_-_5879897 | 2.14 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr20_+_218886 | 2.11 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr5_+_20693724 | 2.01 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr3_-_19899914 | 2.00 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr14_-_5678457 | 1.82 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr7_-_26087807 | 1.81 |
ENSDART00000052989
|
ache
|
acetylcholinesterase |
| chr3_+_1182315 | 1.77 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr15_-_26552393 | 1.75 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr5_-_36328688 | 1.74 |
ENSDART00000011399
|
efnb1
|
ephrin-B1 |
| chr20_+_10545514 | 1.72 |
ENSDART00000153667
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr14_+_34514336 | 1.72 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr14_+_6159356 | 1.68 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr17_+_7595356 | 1.67 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr10_+_25355308 | 1.64 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr24_-_17029374 | 1.64 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr7_+_30875273 | 1.64 |
ENSDART00000173693
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr16_-_42013858 | 1.63 |
ENSDART00000045403
|
etv2
|
ets variant 2 |
| chr7_-_38658411 | 1.57 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr1_-_44940830 | 1.52 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr8_-_52715911 | 1.52 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr5_+_51594209 | 1.51 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr4_-_16345227 | 1.51 |
ENSDART00000079521
|
kera
|
keratocan |
| chr12_-_3940768 | 1.50 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr10_+_37927100 | 1.50 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr19_-_9712530 | 1.49 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr21_-_28901095 | 1.49 |
ENSDART00000180820
|
cxxc5a
|
CXXC finger protein 5a |
| chr5_+_56268436 | 1.47 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr2_+_32780138 | 1.47 |
ENSDART00000082250
|
zgc:136930
|
zgc:136930 |
| chr19_-_103289 | 1.47 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr14_+_6159162 | 1.45 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr3_+_26081343 | 1.44 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr15_+_37105986 | 1.43 |
ENSDART00000157762
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr5_-_35301800 | 1.41 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr16_-_43025885 | 1.39 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr14_-_32016615 | 1.39 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr20_+_34320635 | 1.38 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr15_-_2657508 | 1.35 |
ENSDART00000102086
|
cldna
|
claudin a |
| chr22_+_38194151 | 1.35 |
ENSDART00000121965
|
cp
|
ceruloplasmin |
| chr23_-_19953089 | 1.34 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr24_-_37877743 | 1.34 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr1_+_45080897 | 1.34 |
ENSDART00000129819
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr11_+_11230121 | 1.32 |
ENSDART00000172438
|
myom2a
|
myomesin 2a |
| chr2_-_20599315 | 1.32 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr15_-_21877726 | 1.31 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr7_-_28696556 | 1.28 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr13_+_42124566 | 1.27 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr7_-_28148310 | 1.27 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr23_+_36087219 | 1.25 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr19_+_24488403 | 1.22 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr22_+_661505 | 1.22 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr19_-_12145765 | 1.22 |
ENSDART00000032474
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr15_-_26552652 | 1.21 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr18_+_17583479 | 1.21 |
ENSDART00000186977
ENSDART00000010998 |
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr3_+_39568290 | 1.20 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr8_+_23213320 | 1.20 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr12_-_48477031 | 1.20 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr11_-_10770053 | 1.19 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr12_+_7865470 | 1.19 |
ENSDART00000161683
|
BX548028.1
|
|
| chr16_-_29702447 | 1.18 |
ENSDART00000150617
|
tnfaip8l2b
|
tumor necrosis factor, alpha-induced protein 8-like 2b |
| chr14_-_9199968 | 1.18 |
ENSDART00000146113
|
arhgef9b
|
Cdc42 guanine nucleotide exchange factor (GEF) 9b |
| chr2_-_58201173 | 1.18 |
ENSDART00000166282
|
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr14_-_40389699 | 1.16 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr10_-_27197044 | 1.15 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr7_-_25895189 | 1.15 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr23_+_42338325 | 1.15 |
ENSDART00000169660
|
cyp2aa7
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 |
| chr4_-_22671469 | 1.14 |
ENSDART00000050753
|
cd36
|
CD36 molecule (thrombospondin receptor) |
| chr5_-_41834999 | 1.14 |
ENSDART00000135772
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr23_-_24488696 | 1.13 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr14_-_21064199 | 1.13 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr4_+_25627147 | 1.13 |
ENSDART00000041965
|
acot15
|
acyl-CoA thioesterase 15 |
| chr19_-_32487469 | 1.12 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr19_+_47311020 | 1.12 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr12_-_30583668 | 1.12 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr11_+_25259058 | 1.11 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr21_+_26071874 | 1.09 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr22_+_15624371 | 1.05 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr21_-_17956739 | 1.04 |
ENSDART00000148154
|
stx2a
|
syntaxin 2a |
| chr8_+_45003659 | 1.04 |
ENSDART00000132663
|
si:ch211-163b2.4
|
si:ch211-163b2.4 |
| chr1_+_50987535 | 1.04 |
ENSDART00000140657
|
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr12_+_8168272 | 1.04 |
ENSDART00000054092
|
arid5b
|
AT-rich interaction domain 5B |
| chr4_+_14360372 | 1.03 |
ENSDART00000007103
|
nuak1a
|
NUAK family, SNF1-like kinase, 1a |
| chr22_+_661711 | 1.03 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr22_-_36876133 | 1.02 |
ENSDART00000147006
|
kng1
|
kininogen 1 |
| chr18_-_8885792 | 1.02 |
ENSDART00000143619
|
si:dkey-95h12.1
|
si:dkey-95h12.1 |
| chr7_+_61480296 | 1.02 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr2_-_24289641 | 1.01 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr19_-_12145390 | 1.00 |
ENSDART00000143087
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr9_-_1986014 | 0.99 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr23_+_25708787 | 0.99 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr21_-_17956416 | 0.98 |
ENSDART00000026737
|
stx2a
|
syntaxin 2a |
| chr20_+_54738210 | 0.98 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr18_+_48423973 | 0.98 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr21_-_20341836 | 0.98 |
ENSDART00000176689
|
rbp4l
|
retinol binding protein 4, like |
| chr21_+_17956856 | 0.97 |
ENSDART00000080431
|
dnai1.2
|
dynein, axonemal, intermediate chain 1, paralog 2 |
| chr16_+_55059026 | 0.96 |
ENSDART00000109391
|
LO017815.1
|
Danio rerio nuclear receptor coactivator 7-like (LOC792958), mRNA. |
| chr3_-_40301467 | 0.95 |
ENSDART00000055186
|
atp5mf
|
ATP synthase membrane subunit f |
| chr4_+_77021784 | 0.94 |
ENSDART00000135345
ENSDART00000133855 |
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr7_-_24204665 | 0.93 |
ENSDART00000167141
|
gmpr2
|
guanosine monophosphate reductase 2 |
| chr1_+_54911458 | 0.93 |
ENSDART00000089603
|
golga7ba
|
golgin A7 family, member Ba |
| chr20_-_9760424 | 0.93 |
ENSDART00000104936
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr24_-_17023392 | 0.92 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr15_-_20916251 | 0.91 |
ENSDART00000134053
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr6_+_22337081 | 0.91 |
ENSDART00000128047
ENSDART00000138930 |
uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr23_-_5683147 | 0.90 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr2_-_27900518 | 0.90 |
ENSDART00000109561
ENSDART00000077720 |
zgc:163121
|
zgc:163121 |
| chr6_+_25257728 | 0.90 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr6_+_8176486 | 0.90 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr22_+_8313513 | 0.90 |
ENSDART00000181169
ENSDART00000103911 |
CABZ01077217.1
|
|
| chr21_-_16114061 | 0.88 |
ENSDART00000035742
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr1_+_44941031 | 0.88 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr13_-_2215213 | 0.87 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr20_+_26095530 | 0.86 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr3_+_43460696 | 0.86 |
ENSDART00000164581
|
galr2b
|
galanin receptor 2b |
| chr2_-_5199431 | 0.86 |
ENSDART00000063384
|
phb2a
|
prohibitin 2a |
| chr25_-_10564721 | 0.86 |
ENSDART00000154776
|
galn
|
galanin/GMAP prepropeptide |
| chr10_+_22775253 | 0.85 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr7_-_33351485 | 0.85 |
ENSDART00000146420
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr22_+_5478353 | 0.85 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr18_-_24996634 | 0.85 |
ENSDART00000170210
|
si:ch211-196l7.4
|
si:ch211-196l7.4 |
| chr14_-_12253309 | 0.84 |
ENSDART00000115101
|
myot
|
myotilin |
| chr10_-_42685512 | 0.84 |
ENSDART00000081347
|
stc1l
|
stanniocalcin 1, like |
| chr6_+_7250824 | 0.84 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr9_+_33145522 | 0.83 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr13_-_13294847 | 0.83 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr19_+_18799319 | 0.83 |
ENSDART00000171843
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr8_+_10304981 | 0.82 |
ENSDART00000160766
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr22_+_9472814 | 0.82 |
ENSDART00000112125
ENSDART00000138850 |
cacna2d2b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2b |
| chr10_-_16065185 | 0.81 |
ENSDART00000187266
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr15_+_20403903 | 0.80 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr20_+_40237441 | 0.79 |
ENSDART00000168928
|
si:ch211-199i15.5
|
si:ch211-199i15.5 |
| chr3_-_32859335 | 0.78 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr25_-_21816269 | 0.78 |
ENSDART00000152014
|
zgc:158222
|
zgc:158222 |
| chr5_+_38276582 | 0.78 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr21_-_20342096 | 0.78 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr5_-_32445835 | 0.78 |
ENSDART00000170919
|
ncs1a
|
neuronal calcium sensor 1a |
| chr7_-_24204200 | 0.78 |
ENSDART00000087298
|
gmpr2
|
guanosine monophosphate reductase 2 |
| chr21_-_26071142 | 0.77 |
ENSDART00000004740
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr9_+_22003942 | 0.76 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr10_+_42374770 | 0.76 |
ENSDART00000020000
|
COX5B
|
zgc:86599 |
| chr4_-_5247335 | 0.74 |
ENSDART00000050221
|
atp6v1e1b
|
ATPase H+ transporting V1 subunit E1b |
| chr14_-_29826659 | 0.74 |
ENSDART00000138413
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr24_+_26432541 | 0.74 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr3_-_21280373 | 0.74 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr5_-_63109232 | 0.74 |
ENSDART00000115128
|
usp2b
|
ubiquitin specific peptidase 2b |
| chr4_-_23839789 | 0.73 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr7_+_20467549 | 0.73 |
ENSDART00000173724
|
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr16_+_32559821 | 0.73 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr6_+_41099787 | 0.72 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr12_+_39203745 | 0.72 |
ENSDART00000153661
|
si:dkeyp-106c3.2
|
si:dkeyp-106c3.2 |
| chr9_+_4429593 | 0.72 |
ENSDART00000184855
|
FP015810.1
|
|
| chr6_-_28980756 | 0.72 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr7_+_20475788 | 0.72 |
ENSDART00000171155
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr16_+_32749591 | 0.72 |
ENSDART00000136759
|
prdm13
|
PR domain containing 13 |
| chr25_-_13188678 | 0.71 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr12_+_34896956 | 0.71 |
ENSDART00000055415
|
prph2a
|
peripherin 2a (retinal degeneration, slow) |
| chr18_-_26101800 | 0.70 |
ENSDART00000004692
|
idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr19_-_42945965 | 0.70 |
ENSDART00000142858
|
dclk3
|
doublecortin-like kinase 3 |
| chr6_+_48618512 | 0.69 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr23_+_18722915 | 0.69 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr8_-_17926814 | 0.69 |
ENSDART00000147344
|
lhx8b
|
LIM homeobox 8b |
| chr18_-_8312848 | 0.69 |
ENSDART00000092033
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr10_-_32524771 | 0.69 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr23_+_18722715 | 0.68 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr4_+_2620751 | 0.68 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr7_-_12821277 | 0.68 |
ENSDART00000091584
|
zgc:158785
|
zgc:158785 |
| chr22_+_7486867 | 0.68 |
ENSDART00000034586
|
CELA1 (1 of many)
|
zgc:112302 |
| chr19_+_37701450 | 0.68 |
ENSDART00000087694
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr8_-_39903803 | 0.68 |
ENSDART00000012391
|
cabp1a
|
calcium binding protein 1a |
| chr7_+_34688527 | 0.67 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr6_-_50404732 | 0.66 |
ENSDART00000055510
|
romo1
|
reactive oxygen species modulator 1 |
| chr8_+_1769475 | 0.65 |
ENSDART00000079073
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr10_-_29101715 | 0.65 |
ENSDART00000149674
ENSDART00000171194 ENSDART00000192019 |
si:ch211-103f14.3
|
si:ch211-103f14.3 |
| chr17_-_31058900 | 0.65 |
ENSDART00000134998
ENSDART00000104307 ENSDART00000172721 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr25_+_13791627 | 0.65 |
ENSDART00000159278
|
zgc:92873
|
zgc:92873 |
| chr20_+_43942278 | 0.64 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr12_+_15622621 | 0.64 |
ENSDART00000079784
|
plcd3b
|
phospholipase C, delta 3b |
| chr9_-_54344405 | 0.64 |
ENSDART00000182939
|
CT998556.1
|
|
| chr23_-_33750135 | 0.64 |
ENSDART00000187641
|
bin2a
|
bridging integrator 2a |
| chr19_+_14059349 | 0.64 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr9_+_17348745 | 0.64 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr8_+_19514294 | 0.64 |
ENSDART00000170622
|
si:ch73-281k2.5
|
si:ch73-281k2.5 |
| chr11_+_23704410 | 0.64 |
ENSDART00000112655
|
nfasca
|
neurofascin homolog (chicken) a |
| chr1_-_5455498 | 0.63 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr23_+_36083529 | 0.63 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr20_-_40729364 | 0.62 |
ENSDART00000101014
|
cx32.2
|
connexin 32.2 |
| chr21_-_12119711 | 0.62 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr15_+_1004680 | 0.62 |
ENSDART00000157310
|
si:dkey-77f5.8
|
si:dkey-77f5.8 |
| chr25_-_35599887 | 0.61 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr23_+_6272638 | 0.61 |
ENSDART00000190366
|
syt2a
|
synaptotagmin IIa |
| chr14_+_8343498 | 0.60 |
ENSDART00000164551
|
nrg2b
|
neuregulin 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr5a1a+nr5a2+nr5a5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.6 | 2.3 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.4 | 3.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.4 | 1.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.4 | 1.7 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.4 | 5.2 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.4 | 1.5 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.4 | 1.4 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 1.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 1.5 | GO:2000379 | positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.3 | 0.9 | GO:1901296 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.3 | 0.8 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.3 | 1.3 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.2 | 1.7 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.2 | 2.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.2 | 0.9 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.2 | 1.1 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 1.3 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 1.0 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 0.6 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 1.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.2 | 0.5 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.2 | 0.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 2.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.6 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 1.0 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.8 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.4 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 1.3 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.5 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.1 | 0.5 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.5 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 2.8 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.5 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.5 | GO:1990544 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 1.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 0.5 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 1.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.4 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 1.4 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 0.4 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 1.1 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.8 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 2.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 7.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.8 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.7 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.5 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.5 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 1.0 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 0.3 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 2.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.3 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 2.0 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.5 | GO:0060114 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.7 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 2.7 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 1.1 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.3 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.4 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 2.0 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.3 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.4 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.8 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.6 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.4 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 1.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 1.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.8 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.2 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.7 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.6 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 1.2 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.9 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 1.5 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 2.1 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.9 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.4 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 2.6 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.7 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 1.7 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 0.7 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 1.6 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.0 | GO:0006821 | chloride transport(GO:0006821) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.4 | 2.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.4 | 1.4 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 1.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 2.1 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 3.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 2.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.7 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.0 | GO:0045259 | proton-transporting ATP synthase complex(GO:0045259) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 3.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.7 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.8 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 1.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0098803 | respiratory chain complex(GO:0098803) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 4.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.0 | 0.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 23.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.4 | 5.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 1.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.3 | 1.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.3 | 1.8 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.3 | 1.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.3 | 1.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.3 | 0.8 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 1.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 1.8 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 0.7 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 0.9 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.2 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 1.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 1.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 1.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 2.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.9 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.4 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.5 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 2.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.3 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 1.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.4 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.9 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.7 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.4 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 4.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.7 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 1.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.5 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.8 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.4 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.6 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 4.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 5.4 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.6 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 1.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 4.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 2.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 1.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 3.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 3.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 3.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 5.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.6 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 2.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.8 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |