Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
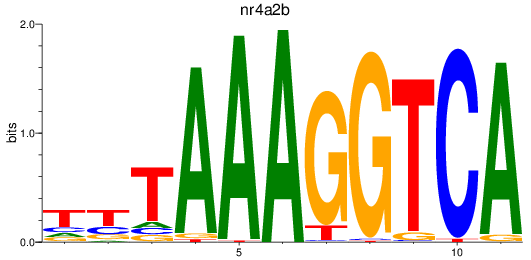
Results for nr4a2b
Z-value: 1.40
Transcription factors associated with nr4a2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr4a2b
|
ENSDARG00000044532 | nuclear receptor subfamily 4, group A, member 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr4a2b | dr11_v1_chr6_+_12462079_12462079 | 0.42 | 2.6e-01 | Click! |
Activity profile of nr4a2b motif
Sorted Z-values of nr4a2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_19332837 | 3.67 |
ENSDART00000078594
|
tyrp1b
|
tyrosinase-related protein 1b |
| chr10_+_9561066 | 3.34 |
ENSDART00000136281
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr20_-_10120442 | 3.29 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr17_+_7595356 | 3.12 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr19_-_5332784 | 2.99 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr5_+_1278092 | 2.87 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr16_+_23972126 | 2.70 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr3_+_23687909 | 2.53 |
ENSDART00000046638
|
hoxb8a
|
homeobox B8a |
| chr7_+_25913225 | 2.48 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr23_-_24488696 | 2.35 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr1_+_16127825 | 2.12 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr20_+_40150612 | 2.07 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr25_+_18964782 | 2.01 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr23_+_36063599 | 2.01 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr5_-_41831646 | 1.97 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr8_+_39634114 | 1.97 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr14_-_17121676 | 1.89 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr3_-_46818001 | 1.88 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr11_+_11201096 | 1.88 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr9_+_31795343 | 1.86 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr3_-_46817838 | 1.84 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr6_-_39764995 | 1.82 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr23_+_27068225 | 1.81 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr7_-_24046999 | 1.74 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr8_+_13106760 | 1.58 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr7_-_19146925 | 1.49 |
ENSDART00000142924
ENSDART00000009695 |
kirrel1a
|
kirre like nephrin family adhesion molecule 1a |
| chr4_+_16795688 | 1.37 |
ENSDART00000132337
|
spx
|
spexin hormone |
| chr4_-_4932619 | 1.35 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr15_-_16884912 | 1.30 |
ENSDART00000062135
|
zgc:103681
|
zgc:103681 |
| chr5_+_37087583 | 1.27 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr9_-_21067971 | 1.27 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr6_-_39765546 | 1.27 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr3_+_23710839 | 1.27 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr14_+_33882973 | 1.25 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr3_-_22212764 | 1.23 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr3_-_28258462 | 1.23 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr15_-_21239416 | 1.23 |
ENSDART00000155787
|
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr6_-_10835849 | 1.23 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr6_-_35738836 | 1.22 |
ENSDART00000111642
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr7_+_31871830 | 1.21 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr17_-_12389259 | 1.17 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr3_+_32553714 | 1.09 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr11_+_2172335 | 1.07 |
ENSDART00000170593
|
hoxc12b
|
homeobox C12b |
| chr5_-_41838354 | 1.06 |
ENSDART00000146793
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr25_-_16969922 | 1.05 |
ENSDART00000111158
|
tigara
|
tp53-induced glycolysis and apoptosis regulator a |
| chr6_-_21189295 | 1.02 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr1_-_51734524 | 1.00 |
ENSDART00000109640
ENSDART00000122628 |
junba
|
JunB proto-oncogene, AP-1 transcription factor subunit a |
| chr23_-_45504991 | 0.98 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr2_+_6885852 | 0.98 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr12_+_36891483 | 0.94 |
ENSDART00000048927
|
cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr6_+_40775800 | 0.87 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr24_+_25471196 | 0.86 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr8_-_7440364 | 0.84 |
ENSDART00000180659
|
hdac6
|
histone deacetylase 6 |
| chr10_-_20524592 | 0.84 |
ENSDART00000185048
|
ddhd2
|
DDHD domain containing 2 |
| chr18_+_13182528 | 0.80 |
ENSDART00000166298
|
zgc:56622
|
zgc:56622 |
| chr1_-_30689004 | 0.77 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr8_-_23578660 | 0.77 |
ENSDART00000039080
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr5_-_16983336 | 0.75 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr12_-_26423439 | 0.75 |
ENSDART00000113978
|
synpo2lb
|
synaptopodin 2-like b |
| chr10_-_29892486 | 0.75 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr7_+_20524064 | 0.75 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr18_-_15771551 | 0.74 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr15_+_29292154 | 0.72 |
ENSDART00000137817
|
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr1_+_31638274 | 0.71 |
ENSDART00000057885
|
fgf8b
|
fibroblast growth factor 8 b |
| chr15_+_20403903 | 0.70 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr19_-_34927201 | 0.70 |
ENSDART00000076518
|
sla1
|
Src-like-adaptor 1 |
| chr11_+_1409622 | 0.65 |
ENSDART00000157042
|
ppp1r3db
|
protein phosphatase 1, regulatory subunit 3Db |
| chr3_+_31177972 | 0.65 |
ENSDART00000185954
|
clec19a
|
C-type lectin domain containing 19A |
| chr5_-_67721368 | 0.65 |
ENSDART00000134386
|
arhgap31
|
Rho GTPase activating protein 31 |
| chr19_-_703898 | 0.64 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr19_+_32158010 | 0.64 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr3_+_24190207 | 0.62 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr10_-_20524387 | 0.62 |
ENSDART00000159060
|
ddhd2
|
DDHD domain containing 2 |
| chr6_-_50404732 | 0.62 |
ENSDART00000055510
|
romo1
|
reactive oxygen species modulator 1 |
| chr6_+_29288223 | 0.62 |
ENSDART00000112099
|
zgc:172121
|
zgc:172121 |
| chr6_-_29288155 | 0.61 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr1_-_20593778 | 0.60 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr23_+_21544227 | 0.59 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr24_-_36802891 | 0.56 |
ENSDART00000088130
|
dcakd
|
dephospho-CoA kinase domain containing |
| chr10_-_11385155 | 0.56 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr16_+_20161805 | 0.55 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr17_+_30545895 | 0.55 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr5_-_41841892 | 0.54 |
ENSDART00000167089
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr8_-_14052349 | 0.53 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr6_-_10320676 | 0.53 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr17_-_17764801 | 0.51 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr3_+_17806213 | 0.49 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr5_-_68093169 | 0.49 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr8_-_42238543 | 0.48 |
ENSDART00000062697
|
gfra2a
|
GDNF family receptor alpha 2a |
| chr20_+_106481 | 0.48 |
ENSDART00000148834
|
si:ch1073-155h21.2
|
si:ch1073-155h21.2 |
| chr14_+_35023923 | 0.47 |
ENSDART00000172171
|
ebf3a
|
early B cell factor 3a |
| chr8_-_979735 | 0.46 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr13_-_41155472 | 0.46 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr7_+_34620418 | 0.45 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr12_-_26415499 | 0.44 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr23_+_26142807 | 0.44 |
ENSDART00000158878
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr7_+_44484853 | 0.43 |
ENSDART00000189079
ENSDART00000121826 |
bean1
|
brain expressed, associated with NEDD4, 1 |
| chr24_+_22022109 | 0.42 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr18_+_31410652 | 0.40 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr13_-_27660955 | 0.40 |
ENSDART00000188651
ENSDART00000134494 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr5_-_66301142 | 0.39 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr6_+_29288006 | 0.38 |
ENSDART00000043496
|
zgc:172121
|
zgc:172121 |
| chr23_-_28141419 | 0.37 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr2_-_48753873 | 0.35 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr21_+_5080789 | 0.33 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr9_+_42066030 | 0.33 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr20_-_1151265 | 0.33 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr11_+_23810148 | 0.32 |
ENSDART00000127846
|
nfasca
|
neurofascin homolog (chicken) a |
| chr22_+_31821815 | 0.31 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr8_-_44004135 | 0.31 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr5_-_63218919 | 0.30 |
ENSDART00000149979
|
tecta
|
tectorin alpha |
| chr7_-_24005268 | 0.30 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr1_+_44826593 | 0.29 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr2_-_32457919 | 0.29 |
ENSDART00000132792
ENSDART00000041319 |
slc4a2a
|
solute carrier family 4 (anion exchanger), member 2a |
| chr18_-_2433011 | 0.28 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr24_+_34089977 | 0.27 |
ENSDART00000157466
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr7_+_38588866 | 0.27 |
ENSDART00000015682
|
ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3, (NADH-coenzyme Q reductase) |
| chr21_-_22724980 | 0.27 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr12_-_47648538 | 0.26 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr2_-_11912347 | 0.25 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr11_-_21363834 | 0.23 |
ENSDART00000080051
|
RASSF5
|
si:dkey-85p17.3 |
| chr23_+_26142613 | 0.21 |
ENSDART00000165046
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr15_+_31526225 | 0.20 |
ENSDART00000154456
|
wdr95
|
WD40 repeat domain 95 |
| chr4_-_18434924 | 0.20 |
ENSDART00000190271
|
socs2
|
suppressor of cytokine signaling 2 |
| chr16_-_35586401 | 0.20 |
ENSDART00000169868
ENSDART00000166606 ENSDART00000162562 |
scmh1
|
Scm polycomb group protein homolog 1 |
| chr14_+_36628131 | 0.19 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr15_+_11840311 | 0.19 |
ENSDART00000167671
|
prkd2
|
protein kinase D2 |
| chr15_+_1705167 | 0.18 |
ENSDART00000081940
|
otol1b
|
otolin 1b |
| chr4_-_18775548 | 0.18 |
ENSDART00000141187
|
def6c
|
differentially expressed in FDCP 6c homolog |
| chr23_+_21638258 | 0.18 |
ENSDART00000104188
|
igsf21b
|
immunoglobin superfamily, member 21b |
| chr13_+_7241170 | 0.17 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr2_-_44971551 | 0.17 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr8_-_8698607 | 0.17 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr12_-_29305220 | 0.17 |
ENSDART00000153458
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr18_+_9382847 | 0.16 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr2_-_3045861 | 0.16 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr5_-_33959868 | 0.13 |
ENSDART00000143652
|
zgc:63972
|
zgc:63972 |
| chr2_-_38282079 | 0.13 |
ENSDART00000145808
|
rnf212b
|
si:ch211-10e2.1 |
| chr18_-_4957022 | 0.12 |
ENSDART00000101686
|
zbbx
|
zinc finger, B-box domain containing |
| chr20_-_45722895 | 0.11 |
ENSDART00000153273
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr7_+_22702437 | 0.11 |
ENSDART00000182054
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr20_-_26937453 | 0.10 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr1_+_54115839 | 0.09 |
ENSDART00000180214
|
LO017722.2
|
|
| chr16_-_16619854 | 0.09 |
ENSDART00000150512
ENSDART00000191306 ENSDART00000181773 ENSDART00000183231 |
cyp21a2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr24_-_24848612 | 0.08 |
ENSDART00000190941
|
crhb
|
corticotropin releasing hormone b |
| chr1_-_53988017 | 0.08 |
ENSDART00000003097
|
RHOU (1 of many)
|
si:ch211-133l11.10 |
| chr3_-_25377163 | 0.07 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr19_+_14059349 | 0.07 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr16_-_22294265 | 0.07 |
ENSDART00000124718
|
aqp10a
|
aquaporin 10a |
| chr8_-_17997845 | 0.06 |
ENSDART00000121660
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr10_+_36354863 | 0.06 |
ENSDART00000159675
|
or104-1
|
odorant receptor, family C, subfamily 104, member 1 |
| chr13_+_21768447 | 0.06 |
ENSDART00000100941
|
chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr10_+_41668483 | 0.05 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr15_-_11341635 | 0.05 |
ENSDART00000055220
|
rab30
|
RAB30, member RAS oncogene family |
| chr8_+_53120278 | 0.05 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr7_+_22702225 | 0.04 |
ENSDART00000173672
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr16_-_16761164 | 0.01 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr21_+_37411547 | 0.00 |
ENSDART00000076320
|
mrps17
|
mitochondrial ribosomal protein S17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr4a2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0050995 | negative regulation of lipid transport(GO:0032369) negative regulation of lipid catabolic process(GO:0050995) |
| 0.7 | 3.7 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.5 | 1.4 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.4 | 1.6 | GO:0030811 | regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) regulation of cofactor metabolic process(GO:0051193) regulation of coenzyme metabolic process(GO:0051196) |
| 0.3 | 4.5 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.3 | 3.1 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 0.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 1.8 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 1.2 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 2.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 2.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.6 | GO:2000379 | protein import into mitochondrial inner membrane(GO:0045039) positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.1 | 1.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.7 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 2.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 3.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 0.7 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.5 | GO:0071422 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.9 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 1.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.7 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 1.0 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 1.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.5 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.1 | 1.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.3 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.5 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.9 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0003272 | endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.0 | 0.7 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 3.1 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 0.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 1.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 1.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.8 | GO:0006476 | protein deacetylation(GO:0006476) |
| 0.0 | 1.5 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.3 | 3.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.3 | 3.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 2.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.2 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 2.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 2.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.9 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 2.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 3.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 6.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.3 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 1.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 3.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.7 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 1.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.8 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.3 | 3.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 1.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.2 | 2.0 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.2 | 0.8 | GO:0042903 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.2 | 2.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.2 | 1.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.8 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 2.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.5 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 1.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 3.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 1.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.7 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.5 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 4.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.1 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 6.3 | GO:0005198 | structural molecule activity(GO:0005198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.1 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |