Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
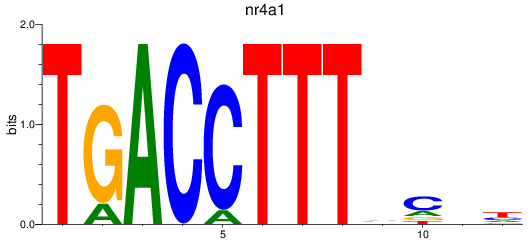
Results for nr4a1
Z-value: 0.82
Transcription factors associated with nr4a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr4a1
|
ENSDARG00000000796 | nuclear receptor subfamily 4, group A, member 1 |
|
nr4a1
|
ENSDARG00000109473 | nuclear receptor subfamily 4, group A, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr4a1 | dr11_v1_chr23_-_32162810_32162810 | 0.95 | 1.1e-04 | Click! |
Activity profile of nr4a1 motif
Sorted Z-values of nr4a1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_29113796 | 1.38 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr7_-_51775688 | 1.29 |
ENSDART00000149793
|
bmp15
|
bone morphogenetic protein 15 |
| chr12_+_21525496 | 1.19 |
ENSDART00000152974
|
ca10a
|
carbonic anhydrase Xa |
| chr12_-_33568174 | 1.17 |
ENSDART00000142329
|
tdrkh
|
tudor and KH domain containing |
| chr18_+_27515640 | 1.16 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr22_-_11833317 | 1.14 |
ENSDART00000125423
ENSDART00000000192 |
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr14_-_7409364 | 1.13 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr5_+_6670945 | 1.07 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr3_-_5829501 | 1.03 |
ENSDART00000091017
|
pkn1b
|
protein kinase N1b |
| chr20_-_32981053 | 1.02 |
ENSDART00000138708
|
nbas
|
neuroblastoma amplified sequence |
| chr11_-_44999858 | 1.01 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr3_+_33745014 | 0.99 |
ENSDART00000159966
|
nacc1a
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing a |
| chr6_-_31348999 | 0.98 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr18_+_44769027 | 0.93 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr14_+_35428152 | 0.88 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr2_-_4787566 | 0.84 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr13_+_43050562 | 0.84 |
ENSDART00000016602
|
cdh23
|
cadherin-related 23 |
| chr13_-_21739142 | 0.83 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr25_+_16214854 | 0.81 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr6_-_41135215 | 0.76 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr1_-_23110740 | 0.75 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr21_-_43527198 | 0.74 |
ENSDART00000126092
|
irs4a
|
insulin receptor substrate 4a |
| chr5_+_19448078 | 0.73 |
ENSDART00000088968
|
ube3b
|
ubiquitin protein ligase E3B |
| chr6_+_515181 | 0.71 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr24_-_26820698 | 0.70 |
ENSDART00000147788
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr20_-_26060154 | 0.70 |
ENSDART00000151950
|
serac1
|
serine active site containing 1 |
| chr23_+_24272421 | 0.69 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr13_-_8888334 | 0.68 |
ENSDART00000059881
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr12_-_6880694 | 0.67 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr13_-_24260609 | 0.67 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr10_-_17745345 | 0.66 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr21_+_10076203 | 0.65 |
ENSDART00000190383
|
CABZ01043506.1
|
|
| chr5_-_14509137 | 0.64 |
ENSDART00000180742
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr1_-_58868306 | 0.62 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr20_-_36917670 | 0.59 |
ENSDART00000046716
|
cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr5_-_3839285 | 0.58 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr4_-_2975461 | 0.58 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr20_+_13883131 | 0.57 |
ENSDART00000003248
ENSDART00000152611 |
nek2
|
NIMA-related kinase 2 |
| chr19_-_3056235 | 0.57 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr19_+_3056450 | 0.56 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr20_-_31743553 | 0.56 |
ENSDART00000087405
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr6_+_30533504 | 0.56 |
ENSDART00000155842
|
wwc3
|
WWC family member 3 |
| chr8_+_21114338 | 0.55 |
ENSDART00000002186
|
uck2a
|
uridine-cytidine kinase 2a |
| chr3_-_27915270 | 0.55 |
ENSDART00000115370
|
mettl22
|
methyltransferase like 22 |
| chr5_+_62611400 | 0.53 |
ENSDART00000132054
|
abr
|
active BCR-related |
| chr3_-_45250924 | 0.52 |
ENSDART00000109017
|
usp31
|
ubiquitin specific peptidase 31 |
| chr6_+_40952031 | 0.52 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr24_-_12770357 | 0.51 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr7_+_17120811 | 0.51 |
ENSDART00000147140
|
prmt3
|
protein arginine methyltransferase 3 |
| chr7_+_34297271 | 0.51 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr25_+_22017182 | 0.51 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr18_-_42333428 | 0.51 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr7_-_31794476 | 0.50 |
ENSDART00000142385
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr10_+_36441124 | 0.50 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr2_+_34767171 | 0.50 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr11_-_31226578 | 0.50 |
ENSDART00000109698
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr20_+_13883353 | 0.50 |
ENSDART00000188006
|
nek2
|
NIMA-related kinase 2 |
| chr7_+_34296789 | 0.49 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_+_44826367 | 0.46 |
ENSDART00000146962
|
STX3
|
zgc:165520 |
| chr17_+_17764979 | 0.45 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr6_-_45995401 | 0.44 |
ENSDART00000154392
|
ca16b
|
carbonic anhydrase XVI b |
| chr5_-_10082244 | 0.44 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr23_+_30730121 | 0.43 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr15_-_6946286 | 0.42 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr14_+_30413312 | 0.42 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr6_-_24392426 | 0.42 |
ENSDART00000163965
|
brdt
|
bromodomain, testis-specific |
| chr8_-_25566347 | 0.42 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr2_-_42827336 | 0.40 |
ENSDART00000140913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr7_+_38667066 | 0.40 |
ENSDART00000013394
|
mtch2
|
mitochondrial carrier homolog 2 |
| chr7_-_42206720 | 0.40 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr10_-_25699454 | 0.40 |
ENSDART00000064376
|
sod1
|
superoxide dismutase 1, soluble |
| chr10_+_7563755 | 0.39 |
ENSDART00000165877
|
purg
|
purine-rich element binding protein G |
| chr7_+_32021669 | 0.39 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr3_-_54500354 | 0.38 |
ENSDART00000124215
|
trip10a
|
thyroid hormone receptor interactor 10a |
| chr16_-_39195318 | 0.38 |
ENSDART00000058546
|
ebag9
|
estrogen receptor binding site associated, antigen, 9 |
| chr11_-_17713987 | 0.37 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr20_+_37825804 | 0.36 |
ENSDART00000152865
|
tatdn3
|
TatD DNase domain containing 3 |
| chr8_+_10823069 | 0.36 |
ENSDART00000081341
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr17_+_15213496 | 0.36 |
ENSDART00000058351
ENSDART00000131663 |
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr25_+_6266009 | 0.35 |
ENSDART00000148995
|
slc25a44a
|
solute carrier family 25, member 44 a |
| chr6_-_40044111 | 0.35 |
ENSDART00000154347
|
si:dkey-197j19.5
|
si:dkey-197j19.5 |
| chr12_+_9817440 | 0.35 |
ENSDART00000137081
ENSDART00000123712 |
rundc3ab
|
RUN domain containing 3Ab |
| chr7_+_65398161 | 0.35 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
| chr15_-_16946124 | 0.34 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr15_+_24676905 | 0.34 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr15_+_28268135 | 0.34 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr2_-_41562868 | 0.34 |
ENSDART00000084597
|
d2hgdh
|
D-2-hydroxyglutarate dehydrogenase |
| chr3_-_41292275 | 0.34 |
ENSDART00000144088
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr7_+_31132588 | 0.33 |
ENSDART00000173702
|
tjp1a
|
tight junction protein 1a |
| chr7_+_30202104 | 0.32 |
ENSDART00000173525
|
wdr76
|
WD repeat domain 76 |
| chr7_-_31794878 | 0.31 |
ENSDART00000099748
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr5_+_23201370 | 0.30 |
ENSDART00000138123
|
tpcn1
|
two pore segment channel 1 |
| chr23_+_20640875 | 0.30 |
ENSDART00000147382
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr1_-_53988017 | 0.30 |
ENSDART00000003097
|
RHOU (1 of many)
|
si:ch211-133l11.10 |
| chr13_-_42673978 | 0.30 |
ENSDART00000133848
ENSDART00000099738 ENSDART00000099729 ENSDART00000169083 |
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr7_+_56253914 | 0.29 |
ENSDART00000073594
|
ankrd11
|
ankyrin repeat domain 11 |
| chr8_-_15991801 | 0.29 |
ENSDART00000132005
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr6_+_3343834 | 0.28 |
ENSDART00000160290
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr8_+_53120278 | 0.28 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr5_-_26330313 | 0.26 |
ENSDART00000148656
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr22_-_16997246 | 0.26 |
ENSDART00000090242
|
nfia
|
nuclear factor I/A |
| chr19_+_19652439 | 0.26 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr24_+_25822999 | 0.26 |
ENSDART00000109809
|
sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr8_+_26083808 | 0.24 |
ENSDART00000099283
|
dalrd3
|
DALR anticodon binding domain containing 3 |
| chr12_-_31457301 | 0.24 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr7_+_42206847 | 0.24 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr10_+_7564106 | 0.24 |
ENSDART00000159042
|
purg
|
purine-rich element binding protein G |
| chr8_+_10404310 | 0.24 |
ENSDART00000185573
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr10_-_11385155 | 0.24 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr8_+_21406769 | 0.23 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr5_+_45677781 | 0.23 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr19_+_5146460 | 0.23 |
ENSDART00000150740
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr13_-_36418921 | 0.23 |
ENSDART00000135804
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr21_+_4320769 | 0.23 |
ENSDART00000168553
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr20_-_31743817 | 0.23 |
ENSDART00000137679
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr11_-_15090118 | 0.22 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr16_-_25680666 | 0.22 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr1_+_44826593 | 0.22 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr5_-_26323137 | 0.22 |
ENSDART00000133823
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr6_+_3282809 | 0.21 |
ENSDART00000187444
ENSDART00000187407 ENSDART00000191883 |
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr17_-_1705013 | 0.21 |
ENSDART00000182864
|
CABZ01086293.1
|
|
| chr16_-_11859309 | 0.21 |
ENSDART00000145754
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr24_-_8214253 | 0.21 |
ENSDART00000160432
|
CABZ01077956.1
|
|
| chr1_+_38153944 | 0.20 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr8_-_15991473 | 0.20 |
ENSDART00000151746
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr8_-_14052349 | 0.19 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr6_-_8498908 | 0.18 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr3_+_32842825 | 0.18 |
ENSDART00000122228
|
prr14
|
proline rich 14 |
| chr6_+_13206516 | 0.18 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr8_-_22542467 | 0.17 |
ENSDART00000182588
ENSDART00000134542 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr11_-_40647190 | 0.17 |
ENSDART00000173217
ENSDART00000173276 ENSDART00000147264 |
fam213b
|
family with sequence similarity 213, member B |
| chr9_-_30346279 | 0.17 |
ENSDART00000089488
|
sytl5
|
synaptotagmin-like 5 |
| chr8_-_45834825 | 0.15 |
ENSDART00000132965
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr12_+_46708920 | 0.15 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr8_-_45835056 | 0.14 |
ENSDART00000022242
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr22_-_16997475 | 0.13 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr8_+_25629941 | 0.12 |
ENSDART00000186925
|
wasa
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) a |
| chr20_+_21391181 | 0.12 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr5_-_23200880 | 0.11 |
ENSDART00000051531
|
iqcd
|
IQ motif containing D |
| chr18_+_31410652 | 0.11 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr24_-_24848612 | 0.10 |
ENSDART00000190941
|
crhb
|
corticotropin releasing hormone b |
| chr3_+_30257582 | 0.08 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr5_-_50600512 | 0.08 |
ENSDART00000190318
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr22_+_21317597 | 0.08 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr9_-_43082945 | 0.07 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr24_-_6628359 | 0.07 |
ENSDART00000169731
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr19_-_40985308 | 0.05 |
ENSDART00000132986
ENSDART00000142360 |
si:ch211-120e1.7
|
si:ch211-120e1.7 |
| chr21_-_25250594 | 0.05 |
ENSDART00000163862
|
nfrkb
|
nuclear factor related to kappaB binding protein |
| chr11_+_33312601 | 0.05 |
ENSDART00000188024
|
cntnap5l
|
contactin associated protein-like 5 like |
| chr5_+_65493699 | 0.05 |
ENSDART00000161567
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr4_-_17391091 | 0.05 |
ENSDART00000056002
|
th2
|
tyrosine hydroxylase 2 |
| chr4_-_77377596 | 0.04 |
ENSDART00000186068
|
slco1e1
|
solute carrier organic anion transporter family, member 1E1 |
| chr19_+_34169055 | 0.04 |
ENSDART00000135592
ENSDART00000186043 |
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr17_+_30546579 | 0.04 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr8_+_20825987 | 0.04 |
ENSDART00000133309
|
si:ch211-133l5.4
|
si:ch211-133l5.4 |
| chr22_+_24673168 | 0.03 |
ENSDART00000135257
|
si:rp71-23d18.8
|
si:rp71-23d18.8 |
| chr6_-_53204808 | 0.03 |
ENSDART00000160955
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr15_+_1705167 | 0.03 |
ENSDART00000081940
|
otol1b
|
otolin 1b |
| chr18_+_50461981 | 0.03 |
ENSDART00000158761
|
CU896640.1
|
|
| chr24_+_2961098 | 0.03 |
ENSDART00000163760
ENSDART00000170835 |
eci2
|
enoyl-CoA delta isomerase 2 |
| chr5_-_19494048 | 0.02 |
ENSDART00000098795
|
mmab
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr20_-_26936887 | 0.02 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr2_+_10127762 | 0.02 |
ENSDART00000100726
|
insl5b
|
insulin-like 5b |
| chr16_-_16619854 | 0.02 |
ENSDART00000150512
ENSDART00000191306 ENSDART00000181773 ENSDART00000183231 |
cyp21a2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr7_+_22792132 | 0.02 |
ENSDART00000135207
ENSDART00000146801 |
rbm4.3
|
RNA binding motif protein 4.3 |
| chr5_+_24156170 | 0.01 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr3_+_24050043 | 0.01 |
ENSDART00000151788
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr25_-_31898552 | 0.01 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr24_-_6024466 | 0.01 |
ENSDART00000040865
|
pdss1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr18_+_9382847 | 0.00 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr10_+_41668483 | 0.00 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr9_-_9419704 | 0.00 |
ENSDART00000138996
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr4a1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.3 | 1.0 | GO:2000622 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 0.9 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.2 | 1.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 1.1 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.2 | 0.8 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 1.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.4 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.8 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.5 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 1.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.4 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.2 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 1.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.6 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.6 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.4 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.5 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 1.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.6 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.7 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.3 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 0.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.3 | 1.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.2 | 0.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 1.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 0.9 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.2 | 1.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 1.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.8 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.4 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.1 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.3 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.8 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.1 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |