Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
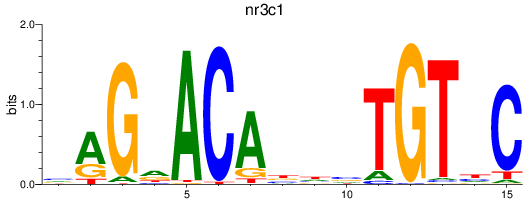
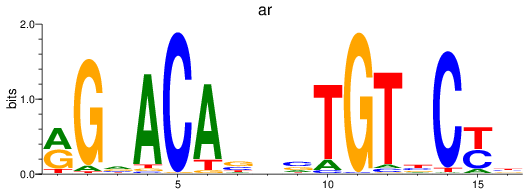
Results for nr3c1_ar
Z-value: 1.41
Transcription factors associated with nr3c1_ar
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr3c1
|
ENSDARG00000025032 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
nr3c1
|
ENSDARG00000112480 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
nr3c1
|
ENSDARG00000116957 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
ar
|
ENSDARG00000067976 | androgen receptor |
|
ar
|
ENSDARG00000114287 | androgen receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ar | dr11_v1_chr5_+_35561607_35561607 | -0.35 | 3.6e-01 | Click! |
| nr3c1 | dr11_v1_chr14_-_23799345_23799349 | -0.25 | 5.2e-01 | Click! |
Activity profile of nr3c1_ar motif
Sorted Z-values of nr3c1_ar motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41096058 | 2.99 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr6_+_41099787 | 2.32 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr12_+_13256415 | 2.11 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr19_+_10855158 | 2.07 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr15_-_9031996 | 1.77 |
ENSDART00000124998
|
rtn2a
|
reticulon 2a |
| chr16_+_50089417 | 1.77 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr22_-_968484 | 1.74 |
ENSDART00000105895
|
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr16_+_10776688 | 1.67 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr16_-_45225520 | 1.55 |
ENSDART00000158855
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr16_+_10777116 | 1.52 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr8_+_7359294 | 1.52 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr2_-_48196092 | 1.50 |
ENSDART00000139944
|
snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr14_-_46228094 | 1.50 |
ENSDART00000172788
|
si:ch211-113d11.5
|
si:ch211-113d11.5 |
| chr7_+_48288762 | 1.49 |
ENSDART00000083569
|
oaz2b
|
ornithine decarboxylase antizyme 2b |
| chr12_-_4683325 | 1.44 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr7_+_18364176 | 1.43 |
ENSDART00000171606
ENSDART00000186368 |
cd248a
|
CD248 molecule, endosialin a |
| chr7_+_58699718 | 1.42 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr7_-_32833153 | 1.41 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr18_-_48745517 | 1.28 |
ENSDART00000097259
|
st3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr20_-_6812688 | 1.27 |
ENSDART00000170934
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr7_+_6969909 | 1.26 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr20_+_25340814 | 1.24 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr4_-_20043484 | 1.19 |
ENSDART00000167780
|
zgc:193726
|
zgc:193726 |
| chr10_-_17988779 | 1.19 |
ENSDART00000132206
ENSDART00000144841 |
si:dkey-242g16.2
|
si:dkey-242g16.2 |
| chr17_-_6730247 | 1.17 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr18_+_16744307 | 1.15 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr4_-_15420452 | 1.12 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr1_-_59176949 | 1.08 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr17_+_33767890 | 1.04 |
ENSDART00000193177
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr21_+_11503212 | 1.01 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr15_-_23647078 | 1.01 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr15_-_23342752 | 1.00 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr9_-_22076368 | 0.99 |
ENSDART00000128486
|
crygm2a
|
crystallin, gamma M2a |
| chr25_+_3358701 | 0.96 |
ENSDART00000104877
|
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr1_+_24076243 | 0.96 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr5_+_32206378 | 0.94 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr23_+_37458602 | 0.93 |
ENSDART00000181686
|
cdaa
|
cytidine deaminase a |
| chr13_+_39236141 | 0.92 |
ENSDART00000111458
|
zgc:172136
|
zgc:172136 |
| chr14_-_28001986 | 0.91 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr20_+_37844035 | 0.87 |
ENSDART00000041397
|
flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr10_-_24343507 | 0.86 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr21_-_26114886 | 0.84 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr23_+_44732863 | 0.82 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr10_+_26667475 | 0.80 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr13_+_912123 | 0.78 |
ENSDART00000169931
|
pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr18_-_24996634 | 0.78 |
ENSDART00000170210
|
si:ch211-196l7.4
|
si:ch211-196l7.4 |
| chr5_-_41494831 | 0.77 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr16_+_32559821 | 0.77 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr19_+_19412692 | 0.77 |
ENSDART00000113580
|
wu:fc38h03
|
wu:fc38h03 |
| chr4_+_4232562 | 0.76 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr20_+_39283849 | 0.75 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr24_+_35564668 | 0.75 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr3_-_32337653 | 0.75 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr15_-_12011390 | 0.73 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr4_-_20156085 | 0.72 |
ENSDART00000055317
|
lrrc17
|
leucine rich repeat containing 17 |
| chr3_-_35602233 | 0.71 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr10_-_36808348 | 0.71 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr20_+_34915945 | 0.71 |
ENSDART00000153064
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr23_+_35708730 | 0.70 |
ENSDART00000009277
|
tuba1a
|
tubulin, alpha 1a |
| chr4_-_11737939 | 0.70 |
ENSDART00000150299
|
podxl
|
podocalyxin-like |
| chr21_-_25565392 | 0.68 |
ENSDART00000144917
ENSDART00000180102 |
si:dkey-17e16.10
|
si:dkey-17e16.10 |
| chr21_-_30648106 | 0.67 |
ENSDART00000160800
ENSDART00000177022 |
phka1b
|
phosphorylase kinase, alpha 1b (muscle) |
| chr16_-_50175069 | 0.66 |
ENSDART00000192979
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr14_-_2355833 | 0.66 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr14_+_22114918 | 0.65 |
ENSDART00000166610
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr5_+_9382301 | 0.65 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr6_-_46875310 | 0.65 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr8_+_35172594 | 0.64 |
ENSDART00000177146
|
BX897670.1
|
|
| chr2_-_155270 | 0.63 |
ENSDART00000131177
|
adcy1b
|
adenylate cyclase 1b |
| chr17_+_6276559 | 0.62 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr5_+_51594209 | 0.62 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr4_-_16354292 | 0.61 |
ENSDART00000139919
|
lum
|
lumican |
| chr8_-_39984593 | 0.61 |
ENSDART00000140127
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr4_+_15968483 | 0.60 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr20_+_33904258 | 0.59 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr21_+_30563115 | 0.59 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr21_-_41873584 | 0.58 |
ENSDART00000188089
|
endou2
|
endonuclease, polyU-specific 2 |
| chr19_-_47452874 | 0.58 |
ENSDART00000025931
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr12_-_43685802 | 0.58 |
ENSDART00000170723
|
zgc:112964
|
zgc:112964 |
| chr4_+_19534833 | 0.57 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr18_+_17428506 | 0.57 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr18_+_8346920 | 0.56 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr19_+_30633453 | 0.55 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr5_-_43935460 | 0.54 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr8_+_19674369 | 0.54 |
ENSDART00000138176
|
foxd2
|
forkhead box D2 |
| chr1_-_39909985 | 0.53 |
ENSDART00000181673
|
stox2a
|
storkhead box 2a |
| chr6_+_9130989 | 0.53 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr18_-_44611252 | 0.53 |
ENSDART00000173095
|
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr10_+_21758811 | 0.53 |
ENSDART00000188827
|
pcdh1g11
|
protocadherin 1 gamma 11 |
| chr11_-_236984 | 0.53 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr12_-_46959990 | 0.52 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr25_-_3470910 | 0.52 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr18_+_17428258 | 0.52 |
ENSDART00000010452
|
zgc:91860
|
zgc:91860 |
| chr9_-_21918963 | 0.52 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr4_+_4803698 | 0.51 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr10_+_34315719 | 0.51 |
ENSDART00000135303
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr17_+_52822422 | 0.51 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr21_+_28478663 | 0.51 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr16_+_46430627 | 0.51 |
ENSDART00000127681
|
rpz6
|
rapunzel 6 |
| chr24_+_7637522 | 0.50 |
ENSDART00000082467
|
cavin1b
|
caveolae associated protein 1b |
| chr24_+_23742690 | 0.50 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr6_-_40583253 | 0.49 |
ENSDART00000032603
|
tspo
|
translocator protein |
| chr11_-_236766 | 0.49 |
ENSDART00000163978
|
dusp7
|
dual specificity phosphatase 7 |
| chr20_-_29475172 | 0.49 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr6_-_6448519 | 0.48 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr19_+_7717449 | 0.48 |
ENSDART00000104719
ENSDART00000146747 |
tuft1b
|
tuftelin 1b |
| chr24_-_9960290 | 0.48 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr22_-_13350240 | 0.48 |
ENSDART00000154095
ENSDART00000155118 |
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr3_-_60571218 | 0.48 |
ENSDART00000178981
|
si:ch73-366l1.5
|
si:ch73-366l1.5 |
| chr25_-_3469576 | 0.47 |
ENSDART00000186738
|
hbp1
|
HMG-box transcription factor 1 |
| chr5_+_23256187 | 0.47 |
ENSDART00000168717
ENSDART00000142915 |
si:dkey-125i10.3
|
si:dkey-125i10.3 |
| chr4_+_25651720 | 0.47 |
ENSDART00000100693
ENSDART00000100717 |
acot16
|
acyl-CoA thioesterase 16 |
| chr12_+_16440708 | 0.47 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr20_-_34801181 | 0.46 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr2_+_13056802 | 0.46 |
ENSDART00000142649
|
prkag2b
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit b |
| chr23_+_4890693 | 0.46 |
ENSDART00000023537
|
tnnc1a
|
troponin C type 1a (slow) |
| chr3_-_58543658 | 0.45 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
| chr5_-_43935119 | 0.45 |
ENSDART00000142271
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr11_-_25829712 | 0.45 |
ENSDART00000103549
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr8_+_1065458 | 0.44 |
ENSDART00000081432
|
sprb
|
sepiapterin reductase b |
| chr20_+_34543365 | 0.44 |
ENSDART00000152073
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr1_-_26782573 | 0.44 |
ENSDART00000090611
|
sh3gl2a
|
SH3 domain containing GRB2 like 2a, endophilin A1 |
| chr19_-_32487469 | 0.44 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr5_-_24238733 | 0.44 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr20_+_46586678 | 0.43 |
ENSDART00000014166
ENSDART00000179266 |
jdp2b
|
Jun dimerization protein 2b |
| chr13_+_25846528 | 0.43 |
ENSDART00000087426
|
bcl11aa
|
B cell CLL/lymphoma 11Aa |
| chr5_-_31901468 | 0.42 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr13_-_45523026 | 0.42 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr5_+_9360394 | 0.42 |
ENSDART00000124642
|
FP236810.2
|
|
| chr23_+_9088191 | 0.41 |
ENSDART00000030811
|
cables2b
|
Cdk5 and Abl enzyme substrate 2b |
| chr17_-_37395460 | 0.40 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr18_+_29156827 | 0.40 |
ENSDART00000137587
ENSDART00000135633 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr2_-_65529 | 0.40 |
ENSDART00000192876
|
zgc:153913
|
zgc:153913 |
| chr25_+_5044780 | 0.40 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr10_+_29431529 | 0.40 |
ENSDART00000158154
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr14_-_2270973 | 0.39 |
ENSDART00000180729
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr7_-_8577190 | 0.39 |
ENSDART00000173174
|
jac3
|
jacalin 3 |
| chr7_-_18168493 | 0.39 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr14_-_36863432 | 0.39 |
ENSDART00000158052
|
rnf130
|
ring finger protein 130 |
| chr8_+_8459192 | 0.38 |
ENSDART00000140942
ENSDART00000014939 |
comta
|
catechol-O-methyltransferase a |
| chr13_+_29510023 | 0.38 |
ENSDART00000187398
|
chst3a
|
carbohydrate (chondroitin 6) sulfotransferase 3a |
| chr5_-_37252111 | 0.38 |
ENSDART00000185110
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr21_+_32338897 | 0.38 |
ENSDART00000110137
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr15_-_12011202 | 0.37 |
ENSDART00000160427
ENSDART00000168715 |
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr2_-_22688651 | 0.36 |
ENSDART00000013863
|
agxtb
|
alanine-glyoxylate aminotransferase b |
| chr17_+_29345606 | 0.36 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr23_+_6752828 | 0.36 |
ENSDART00000105179
|
zgc:158254
|
zgc:158254 |
| chr23_-_39849155 | 0.36 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr1_-_55008882 | 0.36 |
ENSDART00000083572
|
zgc:136864
|
zgc:136864 |
| chr5_-_23362602 | 0.36 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr21_-_19316985 | 0.35 |
ENSDART00000141596
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr25_-_24046870 | 0.35 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr4_-_2014406 | 0.35 |
ENSDART00000180463
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr18_+_50278858 | 0.34 |
ENSDART00000014582
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr4_-_12725513 | 0.34 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr1_-_45049603 | 0.34 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr10_-_43294933 | 0.34 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr6_-_27123327 | 0.34 |
ENSDART00000073881
|
agxta
|
alanine-glyoxylate aminotransferase a |
| chr4_+_76735113 | 0.34 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr15_+_37559570 | 0.33 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr15_-_24883956 | 0.33 |
ENSDART00000113199
|
aipl1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr3_+_62205858 | 0.33 |
ENSDART00000126807
|
zgc:173575
|
zgc:173575 |
| chr11_+_13630107 | 0.33 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr15_-_31000275 | 0.33 |
ENSDART00000132493
ENSDART00000100183 |
lgals9l6
lgals9l5
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 6 lectin, galactoside-binding, soluble, 9 (galectin 9)-like 5 |
| chr24_+_7637264 | 0.32 |
ENSDART00000124409
|
cavin1b
|
caveolae associated protein 1b |
| chr15_-_31372486 | 0.32 |
ENSDART00000127447
|
or111-4
|
odorant receptor, family D, subfamily 111, member 4 |
| chr21_-_131236 | 0.32 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr15_-_29388012 | 0.32 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr4_-_11163112 | 0.31 |
ENSDART00000188854
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr21_-_32289356 | 0.31 |
ENSDART00000183050
|
clk4b
|
CDC-like kinase 4b |
| chr21_+_1378250 | 0.31 |
ENSDART00000186912
|
TCF4
|
transcription factor 4 |
| chr8_-_46700278 | 0.30 |
ENSDART00000143780
|
gpr153
|
G protein-coupled receptor 153 |
| chr23_+_17865953 | 0.30 |
ENSDART00000014723
ENSDART00000140302 ENSDART00000144800 |
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr15_+_7064819 | 0.30 |
ENSDART00000155268
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr19_+_22216778 | 0.30 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr22_+_10010292 | 0.29 |
ENSDART00000180096
|
BX324216.4
|
|
| chr6_-_13114821 | 0.29 |
ENSDART00000164579
ENSDART00000165375 |
zgc:194469
|
zgc:194469 |
| chr6_-_27057702 | 0.29 |
ENSDART00000149363
|
stk25a
|
serine/threonine kinase 25a |
| chr6_-_15096556 | 0.29 |
ENSDART00000185327
|
fhl2b
|
four and a half LIM domains 2b |
| chr15_-_5742531 | 0.29 |
ENSDART00000045985
|
phkg1a
|
phosphorylase kinase, gamma 1a (muscle) |
| chr10_+_41199660 | 0.29 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr16_-_46619967 | 0.29 |
ENSDART00000158341
|
tmem176l.3a
|
transmembrane protein 176l.3a |
| chr11_+_6152643 | 0.29 |
ENSDART00000012789
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr14_+_52571134 | 0.29 |
ENSDART00000166708
|
rpl26
|
ribosomal protein L26 |
| chr5_-_37900350 | 0.28 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr11_-_25829006 | 0.28 |
ENSDART00000159214
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr10_-_24689280 | 0.28 |
ENSDART00000191476
|
si:ch211-287a12.9
|
si:ch211-287a12.9 |
| chr19_-_41213718 | 0.28 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr10_+_26669177 | 0.28 |
ENSDART00000143402
|
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr5_+_32162684 | 0.28 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr19_+_43256986 | 0.28 |
ENSDART00000182336
|
DGKD
|
diacylglycerol kinase delta |
| chr1_-_1631399 | 0.28 |
ENSDART00000176787
|
clic6
|
chloride intracellular channel 6 |
| chr3_-_35800221 | 0.27 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr23_-_5685023 | 0.27 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr20_-_791788 | 0.27 |
ENSDART00000134128
|
impg1a
|
interphotoreceptor matrix proteoglycan 1a |
| chr19_-_205104 | 0.27 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr7_-_35036770 | 0.27 |
ENSDART00000123174
|
galr1b
|
galanin receptor 1b |
| chr7_-_29340427 | 0.26 |
ENSDART00000052584
|
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr5_-_3627110 | 0.26 |
ENSDART00000156071
|
si:zfos-375h5.1
|
si:zfos-375h5.1 |
| chr8_+_53452681 | 0.26 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr3c1_ar
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0051000 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.4 | 1.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.3 | 0.9 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 0.7 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 0.9 | GO:0097037 | heme export(GO:0097037) |
| 0.2 | 4.0 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 0.5 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.2 | 1.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 5.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 0.9 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.4 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 1.6 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.7 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 1.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.4 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 2.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.5 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.7 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 1.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.4 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.2 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 1.7 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.1 | 1.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.3 | GO:0035739 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.0 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.4 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.3 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.1 | 0.3 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.4 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.3 | GO:0060402 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.1 | 1.2 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.2 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.2 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.0 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.4 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.7 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 1.1 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.6 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.6 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0030206 | N-acetylglucosamine metabolic process(GO:0006044) chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.3 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.4 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.1 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 1.0 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 3.9 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.3 | GO:0042102 | positive regulation of T cell proliferation(GO:0042102) |
| 0.0 | 0.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import(GO:0097369) inorganic cation import into cell(GO:0098659) sodium ion import across plasma membrane(GO:0098719) inorganic ion import into cell(GO:0099587) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.1 | GO:0050957 | neuromuscular process controlling balance(GO:0050885) equilibrioception(GO:0050957) |
| 0.0 | 0.5 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.1 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.3 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.1 | 1.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 1.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.6 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 2.1 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 1.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 2.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.4 | 1.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 3.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 0.8 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.6 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 2.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 1.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 1.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.4 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 1.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.6 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.3 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 0.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 2.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.0 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.2 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.3 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.3 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 1.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 1.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.5 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.1 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |