Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
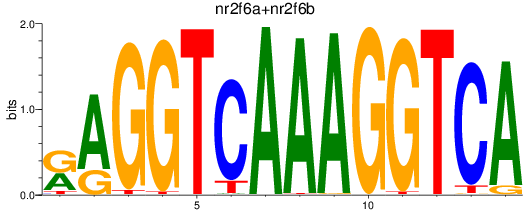
Results for nr2f6a+nr2f6b
Z-value: 0.50
Transcription factors associated with nr2f6a+nr2f6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f6b
|
ENSDARG00000003165 | nuclear receptor subfamily 2, group F, member 6b |
|
nr2f6a
|
ENSDARG00000003607 | nuclear receptor subfamily 2, group F, member 6a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f6b | dr11_v1_chr11_+_6116096_6116096 | 0.70 | 3.7e-02 | Click! |
| nr2f6a | dr11_v1_chr2_+_24374305_24374305 | 0.47 | 2.0e-01 | Click! |
Activity profile of nr2f6a+nr2f6b motif
Sorted Z-values of nr2f6a+nr2f6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_12031958 | 1.37 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr13_-_39159810 | 1.28 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr13_-_39160018 | 1.15 |
ENSDART00000168795
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr8_+_999421 | 1.00 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr4_-_16451375 | 0.98 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr1_+_25848231 | 0.96 |
ENSDART00000027973
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr21_-_44512893 | 0.89 |
ENSDART00000166853
|
zgc:136410
|
zgc:136410 |
| chr2_+_42191592 | 0.79 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr6_+_1787160 | 0.67 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr9_+_54644626 | 0.67 |
ENSDART00000190609
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr11_-_6974022 | 0.65 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr22_-_15587360 | 0.64 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr20_+_2281933 | 0.58 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr5_+_49744713 | 0.57 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr10_-_44027391 | 0.57 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr13_-_37127970 | 0.53 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr11_+_30729745 | 0.51 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr6_-_13783604 | 0.51 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr11_+_11201096 | 0.49 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr23_-_45504991 | 0.47 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr25_+_15647993 | 0.45 |
ENSDART00000186578
ENSDART00000031828 |
spon1b
|
spondin 1b |
| chr1_+_23783349 | 0.45 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr16_-_50229193 | 0.44 |
ENSDART00000161782
ENSDART00000010081 |
etfb
|
electron-transfer-flavoprotein, beta polypeptide |
| chr6_+_60055168 | 0.44 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr6_+_40775800 | 0.43 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr7_-_24046999 | 0.40 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr10_+_20128267 | 0.37 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr7_+_31871830 | 0.30 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr16_+_54875530 | 0.29 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr17_+_450956 | 0.27 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr3_-_3496738 | 0.26 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr24_+_81527 | 0.23 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr23_+_27912079 | 0.23 |
ENSDART00000171859
|
BX539336.1
|
|
| chr18_-_7400075 | 0.19 |
ENSDART00000101250
|
si:dkey-30c15.13
|
si:dkey-30c15.13 |
| chr19_+_2275019 | 0.18 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr18_-_1185772 | 0.18 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr23_+_44634187 | 0.17 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr24_-_21172122 | 0.16 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr23_-_46126444 | 0.15 |
ENSDART00000030004
|
CABZ01071903.1
|
|
| chr1_-_51720633 | 0.14 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr18_-_7399767 | 0.14 |
ENSDART00000181689
|
si:dkey-30c15.13
|
si:dkey-30c15.13 |
| chr9_+_32860345 | 0.13 |
ENSDART00000121751
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr16_-_24605969 | 0.13 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr18_-_18850302 | 0.12 |
ENSDART00000131965
ENSDART00000167624 |
tgm2l
|
transglutaminase 2, like |
| chr3_+_25999477 | 0.11 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr3_+_32553714 | 0.10 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr23_+_44633858 | 0.10 |
ENSDART00000180728
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr13_+_45980163 | 0.10 |
ENSDART00000074547
ENSDART00000005195 |
bsdc1
|
BSD domain containing 1 |
| chr18_-_48550426 | 0.09 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr22_-_16997475 | 0.08 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr18_+_31410652 | 0.08 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr23_+_36340520 | 0.07 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr8_-_979735 | 0.07 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr23_-_21797517 | 0.07 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr5_+_43870389 | 0.07 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr23_+_42810055 | 0.07 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr22_-_16997887 | 0.06 |
ENSDART00000138233
|
nfia
|
nuclear factor I/A |
| chr8_-_8698607 | 0.06 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr18_-_15771551 | 0.05 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr8_+_36803415 | 0.03 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr3_+_39566999 | 0.02 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr6_-_49873020 | 0.01 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr22_-_16997246 | 0.00 |
ENSDART00000090242
|
nfia
|
nuclear factor I/A |
| chr25_+_418932 | 0.00 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f6a+nr2f6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.4 | GO:0019626 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 0.2 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.1 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.9 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 2.0 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.7 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 1.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.4 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |