Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
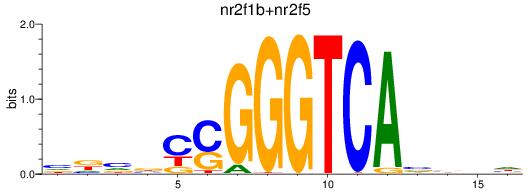
Results for nr2f1b+nr2f5
Z-value: 0.55
Transcription factors associated with nr2f1b+nr2f5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f1b
|
ENSDARG00000017168 | nuclear receptor subfamily 2, group F, member 1b |
|
nr2f5
|
ENSDARG00000033172 | nuclear receptor subfamily 2, group F, member 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f5 | dr11_v1_chr16_+_46294337_46294337 | 0.51 | 1.6e-01 | Click! |
| nr2f1b | dr11_v1_chr10_+_43797130_43797130 | -0.11 | 7.8e-01 | Click! |
Activity profile of nr2f1b+nr2f5 motif
Sorted Z-values of nr2f1b+nr2f5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_37888249 | 0.60 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr7_-_2039060 | 0.49 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr20_-_16906623 | 0.45 |
ENSDART00000012859
ENSDART00000171628 |
psma6b
|
proteasome subunit alpha 6b |
| chr21_+_25688388 | 0.43 |
ENSDART00000125709
|
bicdl2
|
bicaudal-D-related protein 2 |
| chr2_+_52847049 | 0.37 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr13_-_23007813 | 0.33 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr5_+_54400971 | 0.30 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr2_+_58841181 | 0.27 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr4_+_1530287 | 0.25 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr5_+_64856666 | 0.25 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr1_-_55840105 | 0.24 |
ENSDART00000137909
|
samd1b
|
sterile alpha motif domain containing 1b |
| chr22_-_26834043 | 0.23 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr14_-_5817039 | 0.23 |
ENSDART00000131820
|
kazald2
|
Kazal-type serine peptidase inhibitor domain 2 |
| chr15_+_27364394 | 0.21 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr20_-_11178022 | 0.19 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr17_+_389218 | 0.19 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr2_+_16449627 | 0.18 |
ENSDART00000005381
|
zgc:110269
|
zgc:110269 |
| chr17_+_17955063 | 0.17 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr1_-_20824411 | 0.16 |
ENSDART00000182810
|
tll1
|
tolloid-like 1 |
| chr1_+_59154521 | 0.16 |
ENSDART00000130089
ENSDART00000152456 |
soul5l
|
heme-binding protein soul5, like |
| chr2_-_9607879 | 0.16 |
ENSDART00000056899
|
txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr24_+_37700259 | 0.15 |
ENSDART00000066556
|
zmp:0000000624
|
zmp:0000000624 |
| chr2_+_33382648 | 0.15 |
ENSDART00000137207
ENSDART00000098831 |
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr6_+_9677420 | 0.15 |
ENSDART00000064995
|
sumo1
|
small ubiquitin-like modifier 1 |
| chr24_-_21989406 | 0.14 |
ENSDART00000032963
|
apoob
|
apolipoprotein O, b |
| chr8_-_41273768 | 0.14 |
ENSDART00000108518
|
rnf10
|
ring finger protein 10 |
| chr19_+_26828502 | 0.13 |
ENSDART00000183654
ENSDART00000126006 ENSDART00000185341 |
FP085398.1
|
|
| chr3_-_12970418 | 0.13 |
ENSDART00000158747
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr8_-_41273461 | 0.13 |
ENSDART00000190598
|
rnf10
|
ring finger protein 10 |
| chr10_-_29831944 | 0.12 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr3_-_19517462 | 0.12 |
ENSDART00000162027
|
CU571315.2
|
|
| chr14_-_24391424 | 0.12 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr4_-_2440175 | 0.12 |
ENSDART00000058059
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr2_+_43750899 | 0.11 |
ENSDART00000135836
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr23_+_31245395 | 0.11 |
ENSDART00000053588
|
irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr20_+_43925266 | 0.10 |
ENSDART00000037379
|
clic5b
|
chloride intracellular channel 5b |
| chr20_+_25581627 | 0.10 |
ENSDART00000030229
|
cyp2p9
|
cytochrome P450, family 2, subfamily P, polypeptide 9 |
| chr3_+_41917499 | 0.10 |
ENSDART00000028673
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr2_+_21605547 | 0.10 |
ENSDART00000112014
|
mplkip
|
M-phase specific PLK1 interacting protein |
| chr2_+_3696038 | 0.10 |
ENSDART00000150499
ENSDART00000136906 ENSDART00000164152 ENSDART00000128514 ENSDART00000108964 |
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr4_+_76441823 | 0.10 |
ENSDART00000163932
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_+_3980247 | 0.09 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr22_-_31788170 | 0.08 |
ENSDART00000170925
|
pimr207
|
Pim proto-oncogene, serine/threonine kinase, related 207 |
| chr13_+_24834199 | 0.08 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr8_+_23711842 | 0.07 |
ENSDART00000128783
|
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr1_-_26544268 | 0.07 |
ENSDART00000152117
|
si:ch211-202n12.2
|
si:ch211-202n12.2 |
| chr4_+_1722899 | 0.06 |
ENSDART00000021139
|
fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr24_+_42127983 | 0.06 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr11_-_34147205 | 0.06 |
ENSDART00000173216
|
atp13a3
|
ATPase 13A3 |
| chr15_-_34933560 | 0.06 |
ENSDART00000006288
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr18_-_6975175 | 0.06 |
ENSDART00000134194
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr7_+_4474880 | 0.06 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr24_-_21991770 | 0.06 |
ENSDART00000147296
|
si:rp71-1f1.4
|
si:rp71-1f1.4 |
| chr2_+_56213694 | 0.05 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr19_-_48039400 | 0.05 |
ENSDART00000166748
ENSDART00000165921 |
csf3b
|
colony stimulating factor 3 (granulocyte) b |
| chr19_+_5291935 | 0.05 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr1_+_144284 | 0.05 |
ENSDART00000064061
|
prozb
|
protein Z, vitamin K-dependent plasma glycoprotein b |
| chr18_-_27316599 | 0.05 |
ENSDART00000028294
|
zgc:56106
|
zgc:56106 |
| chr8_-_13428740 | 0.04 |
ENSDART00000131826
|
CR354547.3
|
|
| chr24_+_42132962 | 0.04 |
ENSDART00000187739
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr1_-_40132831 | 0.04 |
ENSDART00000135647
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr2_+_2904080 | 0.04 |
ENSDART00000110416
|
eif2b5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon |
| chr18_+_7073130 | 0.03 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr16_+_20895904 | 0.03 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr25_-_10791437 | 0.03 |
ENSDART00000127054
|
BX572619.1
|
|
| chr7_+_27251376 | 0.03 |
ENSDART00000173521
ENSDART00000173962 |
sox6
|
SRY (sex determining region Y)-box 6 |
| chr12_+_7491690 | 0.03 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr19_+_7506380 | 0.02 |
ENSDART00000182837
|
prcc
|
papillary renal cell carcinoma (translocation-associated) |
| chr10_-_21054059 | 0.02 |
ENSDART00000139733
|
pcdh1a
|
protocadherin 1a |
| chr17_+_14711765 | 0.02 |
ENSDART00000012889
|
cx28.6
|
connexin 28.6 |
| chr18_+_3338228 | 0.02 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr2_-_1468258 | 0.02 |
ENSDART00000114431
|
gpaa1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr13_-_50546634 | 0.02 |
ENSDART00000192127
|
CU570781.2
|
|
| chr6_+_103361 | 0.02 |
ENSDART00000151899
|
ldlrb
|
low density lipoprotein receptor b |
| chr8_-_13419049 | 0.02 |
ENSDART00000133656
|
pimr101
|
Pim proto-oncogene, serine/threonine kinase, related 101 |
| chr8_-_13471916 | 0.02 |
ENSDART00000146558
|
pimr105
|
Pim proto-oncogene, serine/threonine kinase, related 105 |
| chr8_-_13486258 | 0.01 |
ENSDART00000137459
|
pimr104
|
Pim proto-oncogene, serine/threonine kinase, related 104 |
| chr7_+_29153018 | 0.01 |
ENSDART00000138065
|
tmppe
|
transmembrane protein with metallophosphoesterase domain |
| chr1_-_47771742 | 0.01 |
ENSDART00000101079
|
neurl1aa
|
neuralized E3 ubiquitin protein ligase 1Aa |
| chr24_+_42117231 | 0.01 |
ENSDART00000067734
|
snx16
|
sorting nexin 16 |
| chr17_+_5351922 | 0.01 |
ENSDART00000105394
|
runx2a
|
runt-related transcription factor 2a |
| chr15_-_41756454 | 0.01 |
ENSDART00000139498
ENSDART00000099302 |
ftr72
|
finTRIM family, member 72 |
| chr8_+_36803415 | 0.01 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr25_+_1549838 | 0.00 |
ENSDART00000115001
|
avpr1aa
|
arginine vasopressin receptor 1Aa |
| chr20_+_53577502 | 0.00 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f1b+nr2f5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.1 | 0.2 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.0 | 0.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.1 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.2 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.3 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |