Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
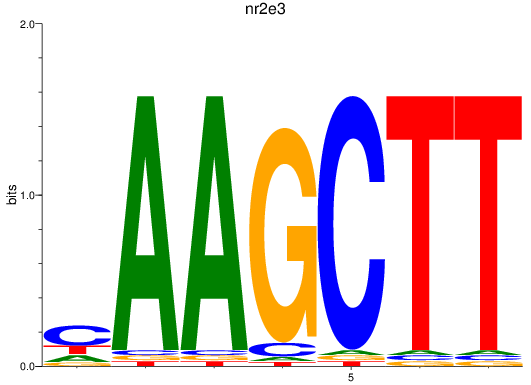
Results for nr2e3
Z-value: 1.21
Transcription factors associated with nr2e3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2e3
|
ENSDARG00000045904 | nuclear receptor subfamily 2, group E, member 3 |
|
nr2e3
|
ENSDARG00000113178 | nuclear receptor subfamily 2, group E, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2e3 | dr11_v1_chr25_+_22274642_22274642 | -0.53 | 1.4e-01 | Click! |
Activity profile of nr2e3 motif
Sorted Z-values of nr2e3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_30293224 | 1.95 |
ENSDART00000101051
|
slbp2
|
stem-loop binding protein 2 |
| chr12_+_30789611 | 1.49 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr19_-_27550768 | 1.49 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr9_-_8314028 | 1.37 |
ENSDART00000102739
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr21_+_15723069 | 1.32 |
ENSDART00000149126
ENSDART00000130628 |
p2rx4a
|
purinergic receptor P2X, ligand-gated ion channel, 4a |
| chr5_+_57924611 | 1.16 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr15_+_38299385 | 1.15 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr20_-_44576949 | 1.12 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr11_-_3494964 | 1.08 |
ENSDART00000162369
|
acap3b
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3b |
| chr17_+_13031497 | 1.07 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr21_-_34261677 | 1.07 |
ENSDART00000124649
ENSDART00000172381 ENSDART00000064320 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr6_-_24384654 | 1.07 |
ENSDART00000164723
|
brdt
|
bromodomain, testis-specific |
| chr19_-_27564458 | 1.06 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr5_+_37747128 | 1.04 |
ENSDART00000148795
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr19_+_8612839 | 1.00 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr10_+_2981090 | 0.97 |
ENSDART00000111830
|
zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr5_+_36768674 | 0.96 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr1_+_16625678 | 0.94 |
ENSDART00000164899
|
pcm1
|
pericentriolar material 1 |
| chr19_+_42231431 | 0.94 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr15_+_38299563 | 0.93 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr4_-_17263210 | 0.93 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr19_-_27564980 | 0.91 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr21_+_4116437 | 0.90 |
ENSDART00000167791
ENSDART00000123759 |
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr10_+_15255012 | 0.89 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr9_-_34269066 | 0.88 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr6_-_25163722 | 0.88 |
ENSDART00000192225
|
znf326
|
zinc finger protein 326 |
| chr23_+_2740741 | 0.88 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr6_+_40992883 | 0.86 |
ENSDART00000076061
|
tgfa
|
transforming growth factor, alpha |
| chr14_-_46617228 | 0.85 |
ENSDART00000161010
ENSDART00000171054 |
prom1a
|
prominin 1a |
| chr8_-_1838315 | 0.85 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr6_+_37754763 | 0.85 |
ENSDART00000110770
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr13_-_6252498 | 0.85 |
ENSDART00000115157
|
tuba4l
|
tubulin, alpha 4 like |
| chr24_-_10014512 | 0.84 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr20_-_20324792 | 0.84 |
ENSDART00000042376
|
snapc1a
|
small nuclear RNA activating complex, polypeptide 1a |
| chr2_+_45382433 | 0.82 |
ENSDART00000142251
|
wdr47a
|
WD repeat domain 47a |
| chr17_+_5931530 | 0.82 |
ENSDART00000168326
ENSDART00000189790 |
znf513b
|
zinc finger protein 513b |
| chr10_+_15255198 | 0.82 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr17_-_2596125 | 0.81 |
ENSDART00000175740
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr13_-_35459928 | 0.81 |
ENSDART00000144109
|
slx4ip
|
SLX4 interacting protein |
| chr22_-_6562618 | 0.81 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr10_+_16911951 | 0.80 |
ENSDART00000164933
|
UNC13B (1 of many)
|
unc-13 homolog B |
| chr22_-_17652914 | 0.80 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr23_+_27756984 | 0.80 |
ENSDART00000137103
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr22_-_17652112 | 0.79 |
ENSDART00000189205
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr24_-_5691956 | 0.79 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr6_+_32338334 | 0.78 |
ENSDART00000065149
|
dock7
|
dedicator of cytokinesis 7 |
| chr2_+_30032303 | 0.77 |
ENSDART00000151841
|
rbm33b
|
RNA binding motif protein 33b |
| chr25_+_15994100 | 0.77 |
ENSDART00000144723
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr19_+_43684376 | 0.77 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr13_-_24877577 | 0.77 |
ENSDART00000182705
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr5_-_20135679 | 0.77 |
ENSDART00000079402
|
usp30
|
ubiquitin specific peptidase 30 |
| chr5_-_62925282 | 0.76 |
ENSDART00000188048
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr18_+_37120096 | 0.76 |
ENSDART00000147385
|
sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr14_+_45028062 | 0.75 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr9_+_33216945 | 0.75 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr8_+_13364950 | 0.75 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr14_+_30328567 | 0.75 |
ENSDART00000105889
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr3_+_24708685 | 0.74 |
ENSDART00000153507
|
mkl1a
|
megakaryoblastic leukemia (translocation) 1a |
| chr24_-_23784701 | 0.73 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr9_+_13641668 | 0.72 |
ENSDART00000135877
|
als2a
|
amyotrophic lateral sclerosis 2a (juvenile) |
| chr13_+_16522608 | 0.72 |
ENSDART00000182838
ENSDART00000143200 |
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr5_-_15494164 | 0.71 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr4_-_77114795 | 0.71 |
ENSDART00000144849
|
CU467646.2
|
|
| chr19_-_5812319 | 0.71 |
ENSDART00000114472
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr5_-_32336613 | 0.71 |
ENSDART00000139732
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr17_-_6954719 | 0.71 |
ENSDART00000188180
|
zbtb24
|
zinc finger and BTB domain containing 24 |
| chr6_-_15641686 | 0.71 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr19_+_41479990 | 0.70 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr4_-_73411863 | 0.68 |
ENSDART00000171434
|
zgc:162958
|
zgc:162958 |
| chr10_-_35149513 | 0.67 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr23_+_43684494 | 0.67 |
ENSDART00000149878
|
otud4
|
OTU deubiquitinase 4 |
| chr14_-_21618005 | 0.66 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr16_-_31598771 | 0.66 |
ENSDART00000016386
|
tg
|
thyroglobulin |
| chr17_-_36860265 | 0.66 |
ENSDART00000182786
ENSDART00000193967 |
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr22_-_22130623 | 0.66 |
ENSDART00000113168
|
CU855878.1
|
|
| chr23_-_22523303 | 0.66 |
ENSDART00000079019
|
spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr20_-_31252809 | 0.66 |
ENSDART00000137236
|
hpcal1
|
hippocalcin-like 1 |
| chr1_-_12397258 | 0.66 |
ENSDART00000144596
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr15_-_33304133 | 0.65 |
ENSDART00000186092
|
nbeab
|
neurobeachin b |
| chr23_+_38957738 | 0.65 |
ENSDART00000193480
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr23_+_12545114 | 0.65 |
ENSDART00000105283
ENSDART00000166990 |
si:zfos-452g4.1
|
si:zfos-452g4.1 |
| chr19_+_42660158 | 0.65 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr25_+_33033633 | 0.65 |
ENSDART00000192336
|
tln2b
|
talin 2b |
| chr24_+_15020402 | 0.65 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr6_+_10333920 | 0.64 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr17_-_2578026 | 0.64 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr20_-_32981053 | 0.64 |
ENSDART00000138708
|
nbas
|
neuroblastoma amplified sequence |
| chr3_+_17456428 | 0.64 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr8_+_29742237 | 0.64 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr12_+_32368574 | 0.64 |
ENSDART00000086389
|
ANKFN1
|
si:ch211-277e21.2 |
| chr15_+_781717 | 0.63 |
ENSDART00000154903
|
znf970
|
zinc finger protein 970 |
| chr20_+_33987465 | 0.63 |
ENSDART00000061751
|
zp3a.2
|
zona pellucida glycoprotein 3a, tandem duplicate 2 |
| chr16_-_17577581 | 0.63 |
ENSDART00000186475
ENSDART00000137215 |
casp2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr23_+_24272421 | 0.62 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr2_-_38114370 | 0.62 |
ENSDART00000131837
|
chd8
|
chromodomain helicase DNA binding protein 8 |
| chr20_+_33991801 | 0.62 |
ENSDART00000061744
|
zp3a.1
|
zona pellucida glycoprotein 3a, tandem duplicate 1 |
| chr25_-_17587785 | 0.62 |
ENSDART00000073679
ENSDART00000146851 |
zgc:66449
|
zgc:66449 |
| chr25_-_7918526 | 0.61 |
ENSDART00000104686
ENSDART00000156761 |
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr24_+_8904135 | 0.61 |
ENSDART00000066782
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr1_+_59314675 | 0.61 |
ENSDART00000161872
ENSDART00000160658 ENSDART00000169792 ENSDART00000160735 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr15_+_14378829 | 0.60 |
ENSDART00000160145
|
si:ch211-11n16.2
|
si:ch211-11n16.2 |
| chr6_-_53885514 | 0.60 |
ENSDART00000173812
ENSDART00000127144 |
cacna2d2a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2a |
| chr15_-_25126842 | 0.60 |
ENSDART00000193670
|
vps53
|
vacuolar protein sorting 53 homolog (S. cerevisiae) |
| chr8_-_25771474 | 0.60 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr9_+_22632126 | 0.59 |
ENSDART00000139434
|
etv5a
|
ets variant 5a |
| chr17_-_6955082 | 0.58 |
ENSDART00000109228
|
zbtb24
|
zinc finger and BTB domain containing 24 |
| chr7_-_15185811 | 0.58 |
ENSDART00000031049
|
si:dkey-172h23.2
|
si:dkey-172h23.2 |
| chr8_-_14080534 | 0.57 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr3_+_32080403 | 0.57 |
ENSDART00000124943
|
aldh16a1
|
aldehyde dehydrogenase 16 family, member A1 |
| chr5_+_33067283 | 0.57 |
ENSDART00000085821
|
npffr2b
|
neuropeptide FF receptor 2b |
| chr13_-_25750910 | 0.57 |
ENSDART00000111567
|
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr9_-_7213772 | 0.57 |
ENSDART00000174720
ENSDART00000092435 |
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr17_-_43594864 | 0.57 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr6_+_12527725 | 0.57 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr9_-_12034444 | 0.57 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr23_-_27505825 | 0.56 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr24_-_21090447 | 0.56 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr9_-_49964810 | 0.56 |
ENSDART00000167098
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr21_-_21178410 | 0.56 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr19_+_37925616 | 0.56 |
ENSDART00000148348
|
nxph1
|
neurexophilin 1 |
| chr9_+_33340311 | 0.56 |
ENSDART00000140064
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr2_+_36114194 | 0.56 |
ENSDART00000113547
|
traj39
|
T-cell receptor alpha joining 39 |
| chr7_-_21905851 | 0.55 |
ENSDART00000111066
ENSDART00000020288 |
epoa
|
erythropoietin a |
| chr3_+_32749613 | 0.55 |
ENSDART00000053684
|
hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase |
| chr16_+_25068576 | 0.55 |
ENSDART00000125838
|
im:7147486
|
im:7147486 |
| chr20_+_29690901 | 0.55 |
ENSDART00000142669
|
mboat2b
|
membrane bound O-acyltransferase domain containing 2b |
| chr19_+_43579786 | 0.55 |
ENSDART00000138404
|
si:ch211-199g17.2
|
si:ch211-199g17.2 |
| chr11_-_15850794 | 0.55 |
ENSDART00000185946
|
rap1ab
|
RAP1A, member of RAS oncogene family b |
| chr4_+_16768961 | 0.54 |
ENSDART00000143926
|
BX649500.1
|
|
| chr10_-_7555326 | 0.54 |
ENSDART00000162191
ENSDART00000186945 |
wrn
|
Werner syndrome |
| chr25_-_7670616 | 0.54 |
ENSDART00000131583
ENSDART00000142794 |
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr22_-_24992532 | 0.54 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr8_-_22274222 | 0.54 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr16_-_12095144 | 0.54 |
ENSDART00000145106
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr12_-_13905307 | 0.53 |
ENSDART00000152400
|
dbf4b
|
DBF4 zinc finger B |
| chr20_+_32523576 | 0.53 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr5_+_31283576 | 0.53 |
ENSDART00000133743
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr6_-_29051773 | 0.53 |
ENSDART00000190508
ENSDART00000180191 ENSDART00000111682 |
evi5b
|
ecotropic viral integration site 5b |
| chr10_+_20392656 | 0.53 |
ENSDART00000160803
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr9_+_24126223 | 0.53 |
ENSDART00000132045
|
pgap1
|
post-GPI attachment to proteins 1 |
| chr9_+_17982737 | 0.52 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr24_-_18809433 | 0.52 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr12_+_13344896 | 0.52 |
ENSDART00000089017
|
rnasen
|
ribonuclease type III, nuclear |
| chr20_-_23426339 | 0.52 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr12_-_4835378 | 0.52 |
ENSDART00000172093
|
si:ch211-93e11.8
|
si:ch211-93e11.8 |
| chr13_-_33207367 | 0.51 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr4_-_14624481 | 0.51 |
ENSDART00000137847
|
plxnb2a
|
plexin b2a |
| chr7_-_40959667 | 0.51 |
ENSDART00000084070
|
rbm33a
|
RNA binding motif protein 33a |
| chr6_+_19948043 | 0.51 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr2_-_22530969 | 0.51 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr7_+_71586485 | 0.51 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr3_-_60589292 | 0.51 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr18_-_6824518 | 0.51 |
ENSDART00000141132
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr2_+_23039041 | 0.51 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr2_-_26604208 | 0.51 |
ENSDART00000133508
ENSDART00000006748 |
yipf1
|
Yip1 domain family, member 1 |
| chr15_+_37936458 | 0.51 |
ENSDART00000154491
|
si:ch73-380l3.2
|
si:ch73-380l3.2 |
| chr6_+_49021703 | 0.51 |
ENSDART00000149394
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr3_-_3439150 | 0.51 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr14_+_21722235 | 0.50 |
ENSDART00000183667
|
stx3a
|
syntaxin 3A |
| chr21_+_37477001 | 0.50 |
ENSDART00000114778
|
amot
|
angiomotin |
| chr16_-_10223741 | 0.50 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr24_+_8904741 | 0.50 |
ENSDART00000140924
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr9_-_32158288 | 0.50 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr11_+_31380495 | 0.50 |
ENSDART00000185073
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr23_+_19564392 | 0.50 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr20_-_39789846 | 0.49 |
ENSDART00000188418
|
rnf217
|
ring finger protein 217 |
| chr13_-_4134141 | 0.49 |
ENSDART00000132354
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr8_-_40442979 | 0.49 |
ENSDART00000157698
ENSDART00000192657 |
tbc1d10aa
|
TBC1 domain family, member 10Aa |
| chr9_+_23770666 | 0.49 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr9_+_8364553 | 0.48 |
ENSDART00000190713
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr2_+_8717155 | 0.48 |
ENSDART00000018114
ENSDART00000137872 ENSDART00000133037 |
usp33
|
ubiquitin specific peptidase 33 |
| chr1_+_47331644 | 0.48 |
ENSDART00000053284
|
bcl9
|
B cell CLL/lymphoma 9 |
| chr10_+_3145707 | 0.47 |
ENSDART00000160046
|
hic2
|
hypermethylated in cancer 2 |
| chr11_-_28911172 | 0.47 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr16_-_8120203 | 0.47 |
ENSDART00000193430
|
snrka
|
SNF related kinase a |
| chr18_-_21746421 | 0.47 |
ENSDART00000188809
|
pskh1
|
protein serine kinase H1 |
| chr7_-_24719655 | 0.46 |
ENSDART00000052805
|
cmtr2
|
cap methyltransferase 2 |
| chr5_-_18474486 | 0.46 |
ENSDART00000090580
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr10_-_35108683 | 0.46 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr2_-_26603958 | 0.46 |
ENSDART00000143085
|
yipf1
|
Yip1 domain family, member 1 |
| chr10_+_585719 | 0.46 |
ENSDART00000180167
|
smad4a
|
SMAD family member 4a |
| chr21_+_10076203 | 0.46 |
ENSDART00000190383
|
CABZ01043506.1
|
|
| chr21_-_14174786 | 0.45 |
ENSDART00000145366
|
whrna
|
whirlin a |
| chr24_-_20321012 | 0.45 |
ENSDART00000163683
|
map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr22_+_34701848 | 0.45 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr25_+_4581214 | 0.45 |
ENSDART00000185552
|
CABZ01068600.1
|
|
| chr5_+_7279104 | 0.45 |
ENSDART00000190014
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr11_+_37803773 | 0.45 |
ENSDART00000019047
|
sox13
|
SRY (sex determining region Y)-box 13 |
| chr25_-_12203952 | 0.45 |
ENSDART00000158204
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr4_+_5848229 | 0.45 |
ENSDART00000161101
ENSDART00000067357 |
lyrm5a
|
LYR motif containing 5a |
| chr8_+_26396552 | 0.44 |
ENSDART00000087151
|
amt
|
aminomethyltransferase |
| chr25_-_7670391 | 0.44 |
ENSDART00000044970
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr4_-_5019113 | 0.44 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr4_-_14191434 | 0.44 |
ENSDART00000142374
ENSDART00000136730 |
pus7l
|
pseudouridylate synthase 7-like |
| chr13_-_36525982 | 0.44 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr23_+_27789795 | 0.44 |
ENSDART00000141458
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr21_+_33311622 | 0.44 |
ENSDART00000163808
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr11_-_27953135 | 0.43 |
ENSDART00000168338
|
ece1
|
endothelin converting enzyme 1 |
| chr13_-_8353515 | 0.43 |
ENSDART00000080351
ENSDART00000190842 |
dhx57
|
DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 |
| chr2_+_50608099 | 0.43 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2e3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.3 | 0.9 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.3 | 1.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 0.5 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 0.9 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 1.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 0.8 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 1.0 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.2 | 0.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 0.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 0.7 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.2 | 0.7 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.2 | 1.5 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.7 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.2 | 0.7 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 0.6 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 2.7 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.1 | 1.3 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.7 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.4 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 1.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.9 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 1.3 | GO:0014074 | response to purine-containing compound(GO:0014074) response to ATP(GO:0033198) response to organophosphorus(GO:0046683) |
| 0.1 | 0.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.4 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.5 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.9 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.5 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.6 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 0.7 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 0.6 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.8 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.3 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 0.3 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 0.4 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 0.5 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.5 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.1 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.6 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.1 | 0.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 1.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.2 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 0.4 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.5 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.7 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.3 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.2 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.2 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.4 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.7 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.9 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:1905133 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.0 | 0.2 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 1.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.6 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.4 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.8 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0021634 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) optic nerve formation(GO:0021634) |
| 0.0 | 0.4 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 1.5 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.4 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.0 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 1.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.6 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 1.3 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.5 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.4 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 1.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.4 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.5 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.3 | GO:0060260 | regulation of transcription initiation from RNA polymerase II promoter(GO:0060260) regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.0 | 0.1 | GO:0061081 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of response to cytokine stimulus(GO:0060760) positive regulation of macrophage cytokine production(GO:0060907) positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) myeloid leukocyte cytokine production(GO:0061082) |
| 0.0 | 0.4 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 3.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.4 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.6 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.6 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.6 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 1.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.7 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.4 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 1.3 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 1.2 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 1.0 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.0 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.5 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.1 | GO:1900044 | regulation of histone ubiquitination(GO:0033182) negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.0 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 1.0 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.1 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.4 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.7 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.8 | GO:0017148 | negative regulation of translation(GO:0017148) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 0.8 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.2 | 0.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 1.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.9 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.6 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 1.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.7 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.4 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.5 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.3 | 1.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 2.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.9 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.2 | 0.6 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 0.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 0.5 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 1.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.9 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.4 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 0.3 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.1 | 1.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.8 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.4 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.3 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 2.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.4 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 1.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.5 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.1 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.2 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.1 | 0.5 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.4 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) TFIIF-class transcription factor binding(GO:0001096) |
| 0.1 | 1.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.7 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.6 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.0 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0008266 | poly-pyrimidine tract binding(GO:0008187) poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0032184 | SUMO binding(GO:0032183) SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 1.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 4.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.6 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.4 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 4.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.6 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.7 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.5 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |