Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
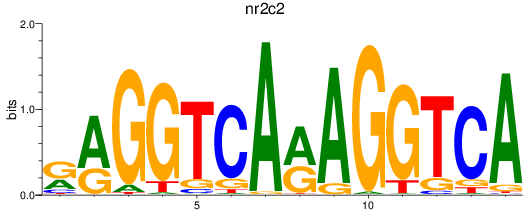
Results for nr2c2
Z-value: 0.90
Transcription factors associated with nr2c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2c2
|
ENSDARG00000042477 | nuclear receptor subfamily 2, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2c2 | dr11_v1_chr8_+_53120278_53120278 | 0.23 | 5.5e-01 | Click! |
Activity profile of nr2c2 motif
Sorted Z-values of nr2c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_44027391 | 1.31 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr5_-_36837846 | 1.12 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr2_-_7666021 | 1.09 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr6_-_13783604 | 1.08 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr11_+_30729745 | 0.95 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr2_+_42191592 | 0.95 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr3_-_31804481 | 0.94 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr12_-_5120339 | 0.93 |
ENSDART00000168759
|
rbp4
|
retinol binding protein 4, plasma |
| chr21_-_19006631 | 0.93 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr12_-_5120175 | 0.90 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr9_+_32978302 | 0.90 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr22_-_23668356 | 0.85 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr13_-_39159810 | 0.84 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr25_-_204019 | 0.80 |
ENSDART00000188440
ENSDART00000191735 |
FP236318.2
|
|
| chr13_-_39160018 | 0.76 |
ENSDART00000168795
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr6_+_60055168 | 0.71 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr12_-_4683325 | 0.69 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr21_-_41870029 | 0.69 |
ENSDART00000182035
|
endou2
|
endonuclease, polyU-specific 2 |
| chr20_+_15015557 | 0.67 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr10_+_35417099 | 0.64 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr10_+_43994471 | 0.62 |
ENSDART00000138242
ENSDART00000186359 |
cldn5b
|
claudin 5b |
| chr4_+_12031958 | 0.61 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr10_-_29900546 | 0.61 |
ENSDART00000147441
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr21_-_35832548 | 0.56 |
ENSDART00000180840
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr25_+_15647993 | 0.56 |
ENSDART00000186578
ENSDART00000031828 |
spon1b
|
spondin 1b |
| chr5_+_36974931 | 0.55 |
ENSDART00000193063
|
gjd1a
|
gap junction protein delta 1a |
| chr22_+_38229321 | 0.54 |
ENSDART00000132670
ENSDART00000104504 |
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr11_+_11201096 | 0.52 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr3_-_12187245 | 0.51 |
ENSDART00000189553
ENSDART00000165131 |
srl
|
sarcalumenin |
| chr2_-_39017838 | 0.50 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr9_+_21151138 | 0.50 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr5_+_38276582 | 0.49 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr25_+_418932 | 0.49 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr10_-_22797959 | 0.49 |
ENSDART00000183269
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr9_-_22834860 | 0.47 |
ENSDART00000146486
|
neb
|
nebulin |
| chr10_-_29272707 | 0.47 |
ENSDART00000022605
ENSDART00000184594 ENSDART00000185371 |
tmem126a
|
transmembrane protein 126A |
| chr22_-_17595310 | 0.46 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr7_+_31871830 | 0.46 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr22_-_15587360 | 0.46 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr3_-_25119839 | 0.45 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr21_+_41743493 | 0.44 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr23_+_44634187 | 0.43 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr6_-_40429411 | 0.43 |
ENSDART00000156005
ENSDART00000156357 |
si:dkey-28n18.9
|
si:dkey-28n18.9 |
| chr8_+_31872992 | 0.41 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr23_-_15216654 | 0.40 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr15_+_19797918 | 0.40 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr19_+_10979983 | 0.40 |
ENSDART00000142975
|
si:ch1073-70f20.1
|
si:ch1073-70f20.1 |
| chr7_+_31838320 | 0.40 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr25_+_20119466 | 0.39 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr19_+_40861853 | 0.39 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr18_-_49283058 | 0.39 |
ENSDART00000076554
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr18_+_31410652 | 0.39 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr8_+_999421 | 0.37 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr11_-_6974022 | 0.36 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr9_+_54644626 | 0.36 |
ENSDART00000190609
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr20_-_21994901 | 0.36 |
ENSDART00000004984
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr19_-_5103141 | 0.36 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr21_-_44512893 | 0.36 |
ENSDART00000166853
|
zgc:136410
|
zgc:136410 |
| chr24_-_33703504 | 0.35 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr19_-_5103313 | 0.35 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr3_-_18410968 | 0.34 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr18_-_48517040 | 0.34 |
ENSDART00000143645
|
kcnj1a.3
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 3 |
| chr18_+_17611627 | 0.33 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr2_-_55853943 | 0.33 |
ENSDART00000122576
|
rx2
|
retinal homeobox gene 2 |
| chr23_+_8797143 | 0.33 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr3_-_18792492 | 0.32 |
ENSDART00000134208
ENSDART00000034373 |
hagh
|
hydroxyacylglutathione hydrolase |
| chr16_-_24612871 | 0.32 |
ENSDART00000155614
ENSDART00000154787 ENSDART00000155983 ENSDART00000156519 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr23_-_45504991 | 0.31 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr9_-_1939232 | 0.31 |
ENSDART00000146131
|
hoxd3a
|
homeobox D3a |
| chr11_+_77526 | 0.30 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr2_-_55861351 | 0.30 |
ENSDART00000059003
|
rx2
|
retinal homeobox gene 2 |
| chr3_-_3496738 | 0.30 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr10_+_20128267 | 0.30 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr7_+_31319876 | 0.30 |
ENSDART00000187611
|
fam189a1
|
family with sequence similarity 189, member A1 |
| chr25_+_18711804 | 0.29 |
ENSDART00000011149
|
fam185a
|
family with sequence similarity 185, member A |
| chr18_-_1185772 | 0.28 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr15_-_6946286 | 0.28 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr18_-_48492951 | 0.28 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr15_-_108414 | 0.28 |
ENSDART00000170044
|
apoa1b
|
apolipoprotein A-Ib |
| chr16_-_39131666 | 0.27 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr18_-_48508585 | 0.27 |
ENSDART00000133364
|
kcnj1a.4
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 4 |
| chr10_+_15340768 | 0.27 |
ENSDART00000046274
ENSDART00000168909 |
trappc13
|
trafficking protein particle complex 13 |
| chr13_-_42724645 | 0.27 |
ENSDART00000046066
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr22_+_9806867 | 0.26 |
ENSDART00000137182
ENSDART00000106103 |
si:ch211-236g6.1
|
si:ch211-236g6.1 |
| chr12_-_35787801 | 0.26 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr6_+_1787160 | 0.26 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr7_-_56180680 | 0.26 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr24_-_21674950 | 0.26 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr23_+_42810055 | 0.25 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr20_-_42702832 | 0.25 |
ENSDART00000134689
ENSDART00000045816 |
plg
|
plasminogen |
| chr4_-_75158035 | 0.24 |
ENSDART00000174353
|
CABZ01066312.1
|
|
| chr7_-_38633867 | 0.24 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr6_-_14004772 | 0.24 |
ENSDART00000185629
|
zgc:92027
|
zgc:92027 |
| chr11_+_24957858 | 0.24 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr6_-_11523987 | 0.24 |
ENSDART00000189363
|
gulp1b
|
GULP, engulfment adaptor PTB domain containing 1b |
| chr10_+_31244619 | 0.24 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr20_+_1996202 | 0.24 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr21_+_8198652 | 0.23 |
ENSDART00000011096
|
nr6a1b
|
nuclear receptor subfamily 6, group A, member 1b |
| chr11_-_27778831 | 0.23 |
ENSDART00000023823
|
cass4
|
Cas scaffolding protein family member 4 |
| chr17_+_1514711 | 0.23 |
ENSDART00000165641
|
akt1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr15_-_29598679 | 0.23 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr7_-_36113098 | 0.23 |
ENSDART00000064913
|
fto
|
fat mass and obesity associated |
| chr17_+_15788100 | 0.23 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr5_-_23362602 | 0.23 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr23_-_637347 | 0.23 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr13_+_28819768 | 0.23 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr3_+_301479 | 0.23 |
ENSDART00000165169
|
CABZ01075611.1
|
|
| chr10_+_21650828 | 0.22 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr17_+_15213496 | 0.22 |
ENSDART00000058351
ENSDART00000131663 |
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr13_-_8692432 | 0.22 |
ENSDART00000058106
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr23_+_27912079 | 0.22 |
ENSDART00000171859
|
BX539336.1
|
|
| chr16_+_7985886 | 0.22 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr23_+_216012 | 0.22 |
ENSDART00000181115
ENSDART00000004678 ENSDART00000190439 ENSDART00000189322 |
PDZD4
|
si:ch73-162j3.4 |
| chr5_-_23317477 | 0.21 |
ENSDART00000090171
|
nlgn3b
|
neuroligin 3b |
| chr13_+_713941 | 0.21 |
ENSDART00000109737
|
cox15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr20_+_15164482 | 0.21 |
ENSDART00000181842
ENSDART00000063877 |
prdx6
|
peroxiredoxin 6 |
| chr22_-_16997475 | 0.21 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr10_-_309894 | 0.21 |
ENSDART00000163287
|
CABZ01049607.1
|
|
| chr9_-_9980704 | 0.21 |
ENSDART00000130243
ENSDART00000193475 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr11_+_25101220 | 0.21 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr13_-_8692860 | 0.20 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr16_-_38518270 | 0.20 |
ENSDART00000131899
|
rspo2
|
R-spondin 2 |
| chr6_+_39930825 | 0.20 |
ENSDART00000182352
ENSDART00000186643 ENSDART00000190513 ENSDART00000065092 ENSDART00000188645 |
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr3_+_27770110 | 0.20 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr7_-_24046999 | 0.20 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr21_+_7605803 | 0.19 |
ENSDART00000121813
|
wdr41
|
WD repeat domain 41 |
| chr6_-_40722200 | 0.19 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr22_-_16997887 | 0.19 |
ENSDART00000138233
|
nfia
|
nuclear factor I/A |
| chr3_+_32553714 | 0.19 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr10_-_322769 | 0.19 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr19_-_24136233 | 0.18 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr12_-_33568174 | 0.18 |
ENSDART00000142329
|
tdrkh
|
tudor and KH domain containing |
| chr23_+_2740741 | 0.18 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr1_-_59240975 | 0.18 |
ENSDART00000166170
|
mvb12a
|
multivesicular body subunit 12A |
| chr2_+_57801960 | 0.18 |
ENSDART00000147966
|
si:dkeyp-68b7.10
|
si:dkeyp-68b7.10 |
| chr1_-_54063520 | 0.18 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr24_-_21913426 | 0.18 |
ENSDART00000081178
|
c1qtnf9
|
C1q and TNF related 9 |
| chr19_+_177455 | 0.18 |
ENSDART00000111580
|
tmem65
|
transmembrane protein 65 |
| chr6_-_40722480 | 0.18 |
ENSDART00000188187
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr17_+_53425037 | 0.17 |
ENSDART00000154983
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr7_+_24115082 | 0.17 |
ENSDART00000182718
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr8_+_22582146 | 0.17 |
ENSDART00000157655
ENSDART00000189892 |
CT583651.2
|
|
| chr9_+_35077546 | 0.17 |
ENSDART00000142688
|
si:dkey-192g7.3
|
si:dkey-192g7.3 |
| chr6_-_39270851 | 0.17 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr16_-_1503023 | 0.17 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr24_-_11446156 | 0.17 |
ENSDART00000143921
ENSDART00000066778 |
acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr11_-_42472941 | 0.17 |
ENSDART00000166624
|
arf4b
|
ADP-ribosylation factor 4b |
| chr9_-_18742704 | 0.17 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr3_-_5829501 | 0.17 |
ENSDART00000091017
|
pkn1b
|
protein kinase N1b |
| chr10_-_38456382 | 0.17 |
ENSDART00000182129
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr20_+_47953047 | 0.17 |
ENSDART00000079734
|
hadhaa
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha a |
| chr12_+_48972915 | 0.17 |
ENSDART00000170695
|
lrit1b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1b |
| chr5_+_22579975 | 0.16 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr3_-_34180364 | 0.16 |
ENSDART00000151819
ENSDART00000003133 |
yipf2
|
Yip1 domain family, member 2 |
| chr3_+_34180835 | 0.16 |
ENSDART00000055252
|
timm29
|
translocase of inner mitochondrial membrane 29 |
| chr18_-_7400075 | 0.16 |
ENSDART00000101250
|
si:dkey-30c15.13
|
si:dkey-30c15.13 |
| chr25_+_28893615 | 0.16 |
ENSDART00000156994
ENSDART00000075151 |
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr20_-_49650293 | 0.16 |
ENSDART00000151239
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr22_-_16997246 | 0.16 |
ENSDART00000090242
|
nfia
|
nuclear factor I/A |
| chr11_+_11974708 | 0.16 |
ENSDART00000125060
|
zgc:64002
|
zgc:64002 |
| chr7_+_59649746 | 0.16 |
ENSDART00000181067
|
rpl34
|
ribosomal protein L34 |
| chr20_-_32270866 | 0.15 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr8_-_25329967 | 0.15 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr23_-_28808480 | 0.15 |
ENSDART00000078167
|
dffa
|
DNA fragmentation factor, alpha polypeptide |
| chr20_+_54738210 | 0.15 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr4_-_858434 | 0.15 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr23_-_45705525 | 0.15 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr8_+_4745340 | 0.15 |
ENSDART00000182894
|
ufd1l
|
ubiquitin recognition factor in ER associated degradation 1 |
| chr1_-_45888608 | 0.15 |
ENSDART00000139219
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr10_-_105100 | 0.15 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr19_-_24484758 | 0.15 |
ENSDART00000052422
|
mtx1b
|
metaxin 1b |
| chr12_-_48992527 | 0.15 |
ENSDART00000169696
|
CDHR1 (1 of many)
|
cadherin related family member 1 |
| chr23_+_19790962 | 0.14 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr8_+_15251448 | 0.14 |
ENSDART00000063717
|
zgc:171480
|
zgc:171480 |
| chr20_+_43379029 | 0.14 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr16_-_24605969 | 0.14 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr2_-_3614005 | 0.14 |
ENSDART00000110399
|
pter
|
phosphotriesterase related |
| chr1_-_53750522 | 0.14 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr1_+_44582369 | 0.14 |
ENSDART00000003022
ENSDART00000137980 |
med19b
|
mediator complex subunit 19b |
| chr2_-_42558549 | 0.14 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr10_+_26834985 | 0.14 |
ENSDART00000147518
|
fam89b
|
family with sequence similarity 89, member B |
| chr4_+_9400012 | 0.14 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr23_-_42810664 | 0.14 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr20_+_2281933 | 0.14 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr21_-_22317920 | 0.14 |
ENSDART00000191083
ENSDART00000108701 |
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr2_-_22660232 | 0.14 |
ENSDART00000174742
|
thap4
|
THAP domain containing 4 |
| chr21_-_5856050 | 0.14 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr25_+_22017182 | 0.14 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr20_-_47953524 | 0.14 |
ENSDART00000167986
|
HADHB
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr19_+_2275019 | 0.13 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr23_-_46126444 | 0.13 |
ENSDART00000030004
|
CABZ01071903.1
|
|
| chr20_+_7243879 | 0.13 |
ENSDART00000122298
|
bsnd
|
barttin CLCNK-type chloride channel accessory beta subunit |
| chr24_+_81527 | 0.13 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr25_+_31929325 | 0.13 |
ENSDART00000181095
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr3_-_32590164 | 0.13 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr14_+_30413758 | 0.13 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr22_-_3595439 | 0.13 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr16_-_21047872 | 0.13 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr11_+_6116503 | 0.13 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr18_-_15771551 | 0.13 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.2 | 0.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.7 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.2 | 0.9 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.6 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) reverse cholesterol transport(GO:0043691) |
| 0.1 | 0.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 1.0 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.3 | GO:0046551 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.2 | GO:0033335 | anal fin development(GO:0033335) |
| 0.1 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.4 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 0.6 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.2 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.1 | 0.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.7 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 0.6 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.1 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.0 | 0.1 | GO:0032369 | negative regulation of lipid transport(GO:0032369) negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.3 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.2 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0006921 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 1.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.3 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 1.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.3 | 1.8 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 0.7 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.7 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.0 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 1.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.2 | GO:1990931 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 0.6 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 3.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.0 | 0.1 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |