Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
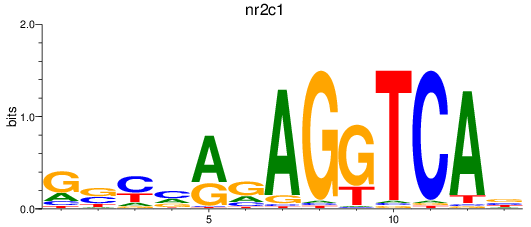
Results for nr2c1
Z-value: 1.60
Transcription factors associated with nr2c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2c1
|
ENSDARG00000045527 | nuclear receptor subfamily 2, group C, member 1 |
|
nr2c1
|
ENSDARG00000111803 | nuclear receptor subfamily 2, group C, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2c1 | dr11_v1_chr4_-_25858620_25858620 | -0.94 | 1.8e-04 | Click! |
Activity profile of nr2c1 motif
Sorted Z-values of nr2c1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_43866526 | 4.29 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr21_+_1119046 | 4.23 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr6_+_154556 | 3.95 |
ENSDART00000193153
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr10_-_21362071 | 3.81 |
ENSDART00000125167
|
avd
|
avidin |
| chr19_-_8798178 | 3.26 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr3_-_34724879 | 2.76 |
ENSDART00000177021
|
thraa
|
thyroid hormone receptor alpha a |
| chr18_+_27515640 | 2.47 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr18_-_43866001 | 2.46 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr5_+_29851433 | 2.42 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr1_+_24387659 | 2.40 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr9_-_32912638 | 2.36 |
ENSDART00000110582
|
fam160a2
|
family with sequence similarity 160, member A2 |
| chr24_+_39137001 | 2.31 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr5_-_16472719 | 2.27 |
ENSDART00000162071
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr20_-_32981053 | 2.26 |
ENSDART00000138708
|
nbas
|
neuroblastoma amplified sequence |
| chr17_-_6613458 | 2.24 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr12_+_48681601 | 2.13 |
ENSDART00000187831
|
uros
|
uroporphyrinogen III synthase |
| chr15_+_47386939 | 2.07 |
ENSDART00000128224
|
FO904873.1
|
|
| chr3_-_58455289 | 1.93 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr8_+_50534948 | 1.91 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr8_-_4097722 | 1.89 |
ENSDART00000135006
|
cux2b
|
cut-like homeobox 2b |
| chr20_-_36671660 | 1.78 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr3_-_34561624 | 1.74 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr19_-_27564458 | 1.72 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr21_-_1972236 | 1.72 |
ENSDART00000192858
ENSDART00000189962 ENSDART00000182461 ENSDART00000165547 |
WDR7
|
WD repeat domain 7 |
| chr23_+_39346774 | 1.71 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr5_+_31049742 | 1.68 |
ENSDART00000173097
|
zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr10_+_36441124 | 1.68 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr18_-_158541 | 1.62 |
ENSDART00000188914
ENSDART00000191052 |
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr23_+_39346930 | 1.58 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr21_+_15883546 | 1.56 |
ENSDART00000186325
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr2_-_4787566 | 1.53 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr3_+_1179601 | 1.51 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr17_+_654759 | 1.49 |
ENSDART00000193703
|
CABZ01079748.1
|
|
| chr22_-_7050 | 1.48 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr21_+_13245302 | 1.45 |
ENSDART00000189498
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr7_+_34296789 | 1.41 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr7_+_34297271 | 1.41 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr3_-_36419641 | 1.39 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr11_+_24314148 | 1.38 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr10_+_15340768 | 1.36 |
ENSDART00000046274
ENSDART00000168909 |
trappc13
|
trafficking protein particle complex 13 |
| chr8_-_12468744 | 1.35 |
ENSDART00000135019
|
FIBCD1 (1 of many)
|
si:dkeyp-51b7.3 |
| chr8_+_10862353 | 1.28 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr14_+_1719367 | 1.27 |
ENSDART00000157696
|
trpc7b
|
transient receptor potential cation channel, subfamily C, member 7b |
| chr2_-_57076687 | 1.26 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr11_-_36020005 | 1.25 |
ENSDART00000031993
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr10_-_40939706 | 1.24 |
ENSDART00000059795
ENSDART00000190510 |
bmp1b
|
bone morphogenetic protein 1b |
| chr7_+_14005111 | 1.23 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr24_-_29586082 | 1.23 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr13_+_7575563 | 1.21 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr1_-_58868306 | 1.20 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr5_-_3839285 | 1.19 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr6_+_40523370 | 1.18 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr4_-_2975461 | 1.18 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr6_+_40952031 | 1.15 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr10_-_38443312 | 1.13 |
ENSDART00000141260
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr19_+_19652439 | 1.10 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr25_-_35113891 | 1.10 |
ENSDART00000190724
|
zgc:165555
|
zgc:165555 |
| chr11_+_24313931 | 1.09 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr11_+_45343245 | 1.09 |
ENSDART00000160904
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr10_-_40939303 | 1.08 |
ENSDART00000134295
|
bmp1b
|
bone morphogenetic protein 1b |
| chr19_-_27564980 | 1.08 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr12_-_22509069 | 1.05 |
ENSDART00000179755
ENSDART00000109707 |
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr3_-_18805225 | 1.03 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr19_+_2631565 | 1.02 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr3_-_33494163 | 1.00 |
ENSDART00000020342
|
sgsm3
|
small G protein signaling modulator 3 |
| chr13_-_36566260 | 0.97 |
ENSDART00000030133
|
synj2bp
|
synaptojanin 2 binding protein |
| chr9_+_38502524 | 0.95 |
ENSDART00000087229
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr23_+_22597624 | 0.93 |
ENSDART00000054337
|
gpr157
|
G protein-coupled receptor 157 |
| chr13_-_30143127 | 0.93 |
ENSDART00000146097
|
tysnd1
|
trypsin domain containing 1 |
| chr25_+_22017182 | 0.92 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr9_+_38503233 | 0.92 |
ENSDART00000140331
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr12_+_26670778 | 0.92 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr12_+_27096835 | 0.92 |
ENSDART00000149475
|
ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr10_-_7555326 | 0.91 |
ENSDART00000162191
ENSDART00000186945 |
wrn
|
Werner syndrome |
| chr10_-_8197049 | 0.91 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr25_-_27665978 | 0.89 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr12_+_23866368 | 0.88 |
ENSDART00000188652
ENSDART00000192478 |
svila
|
supervillin a |
| chr13_+_13805385 | 0.88 |
ENSDART00000166859
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr16_+_30002605 | 0.88 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr19_+_3056450 | 0.87 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr3_+_31680592 | 0.87 |
ENSDART00000172456
|
mylk5
|
myosin, light chain kinase 5 |
| chr21_+_38089036 | 0.86 |
ENSDART00000147219
|
klf8
|
Kruppel-like factor 8 |
| chr14_+_30413312 | 0.84 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr1_+_54737353 | 0.82 |
ENSDART00000130675
ENSDART00000162075 |
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr3_+_40289418 | 0.79 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr10_-_7555660 | 0.77 |
ENSDART00000163689
|
wrn
|
Werner syndrome |
| chr24_-_1021318 | 0.76 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr23_+_2914577 | 0.74 |
ENSDART00000184897
|
DHX35
|
zgc:158828 |
| chr16_-_25680666 | 0.73 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr17_-_49407091 | 0.73 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr11_+_24819324 | 0.72 |
ENSDART00000184549
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr7_+_13988075 | 0.72 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr12_+_31783066 | 0.71 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr15_-_28618502 | 0.71 |
ENSDART00000086902
|
slc6a4a
|
solute carrier family 6 (neurotransmitter transporter), member 4a |
| chr20_-_874807 | 0.70 |
ENSDART00000020506
|
snx14
|
sorting nexin 14 |
| chr23_+_39970425 | 0.69 |
ENSDART00000165376
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr23_+_37482727 | 0.68 |
ENSDART00000162737
|
agmat
|
agmatine ureohydrolase (agmatinase) |
| chr15_-_31213119 | 0.66 |
ENSDART00000182009
|
cuedc1b
|
CUE domain containing 1b |
| chr24_+_14240196 | 0.64 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr10_+_589501 | 0.63 |
ENSDART00000188415
|
LO018557.1
|
|
| chr5_-_26330313 | 0.61 |
ENSDART00000148656
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr11_+_3499111 | 0.60 |
ENSDART00000113096
ENSDART00000171621 |
pusl1
|
pseudouridylate synthase-like 1 |
| chr23_-_45319592 | 0.60 |
ENSDART00000189861
|
ccdc171
|
coiled-coil domain containing 171 |
| chr20_-_29499363 | 0.59 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr22_-_3595439 | 0.58 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr9_+_24936496 | 0.58 |
ENSDART00000157474
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr15_+_28268135 | 0.58 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr11_+_24819624 | 0.57 |
ENSDART00000155514
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr2_-_15040345 | 0.56 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr19_-_24136233 | 0.55 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr22_-_7129631 | 0.54 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr15_+_28119284 | 0.54 |
ENSDART00000188366
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr1_+_58182400 | 0.54 |
ENSDART00000144784
|
si:ch211-15j1.1
|
si:ch211-15j1.1 |
| chr8_-_11146924 | 0.53 |
ENSDART00000126824
|
asb13b
|
ankyrin repeat and SOCS box containing 13b |
| chr5_+_57328535 | 0.52 |
ENSDART00000149646
|
slc31a1
|
solute carrier family 31 (copper transporter), member 1 |
| chr1_-_53750522 | 0.52 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr4_-_7875808 | 0.50 |
ENSDART00000162276
|
nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr3_+_54047342 | 0.50 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr7_-_42206720 | 0.49 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr25_+_27410352 | 0.48 |
ENSDART00000154362
|
pot1
|
protection of telomeres 1 homolog |
| chr11_-_17713987 | 0.48 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr16_-_39131666 | 0.48 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr1_+_59328030 | 0.47 |
ENSDART00000172464
|
CABZ01052576.1
|
|
| chr1_-_44581937 | 0.47 |
ENSDART00000009858
|
tmx2b
|
thioredoxin-related transmembrane protein 2b |
| chr1_+_57311901 | 0.47 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr9_+_40546677 | 0.47 |
ENSDART00000132911
|
vwc2l
|
von Willebrand factor C domain containing protein 2-like |
| chr14_+_989733 | 0.46 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr6_-_39903393 | 0.46 |
ENSDART00000085945
|
tsfm
|
Ts translation elongation factor, mitochondrial |
| chr3_+_26035550 | 0.43 |
ENSDART00000078137
|
ankrd54
|
ankyrin repeat domain 54 |
| chr6_-_51541488 | 0.43 |
ENSDART00000156336
|
si:dkey-6e2.2
|
si:dkey-6e2.2 |
| chr7_+_7495034 | 0.43 |
ENSDART00000102629
ENSDART00000180475 |
nit1
|
nitrilase 1 |
| chr21_-_43171249 | 0.43 |
ENSDART00000021037
ENSDART00000148630 |
hspa4a
|
heat shock protein 4a |
| chr15_+_21711671 | 0.40 |
ENSDART00000136151
|
NKAPD1
|
zgc:162339 |
| chr3_+_27770110 | 0.40 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr5_+_24156170 | 0.38 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr6_-_18393476 | 0.37 |
ENSDART00000168309
|
trim25
|
tripartite motif containing 25 |
| chr7_+_50109239 | 0.37 |
ENSDART00000021605
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr13_+_713941 | 0.37 |
ENSDART00000109737
|
cox15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr19_-_47571456 | 0.37 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr21_+_43171013 | 0.36 |
ENSDART00000151362
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr4_-_965267 | 0.36 |
ENSDART00000093289
|
atp23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr7_-_30553588 | 0.36 |
ENSDART00000139546
|
sltm
|
SAFB-like, transcription modulator |
| chr18_+_38807239 | 0.34 |
ENSDART00000184332
|
fam214a
|
family with sequence similarity 214, member A |
| chr13_-_303137 | 0.33 |
ENSDART00000099131
|
chs1
|
chitin synthase 1 |
| chr4_+_77993103 | 0.33 |
ENSDART00000174392
|
terfa
|
telomeric repeat binding factor a |
| chr13_+_42406883 | 0.33 |
ENSDART00000084354
|
cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr1_-_46859398 | 0.32 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr1_+_58260886 | 0.31 |
ENSDART00000110453
|
si:dkey-222h21.8
|
si:dkey-222h21.8 |
| chr25_+_18587338 | 0.30 |
ENSDART00000180287
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr7_+_42206847 | 0.30 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr4_+_77993977 | 0.29 |
ENSDART00000174118
|
terfa
|
telomeric repeat binding factor a |
| chr1_+_58303892 | 0.28 |
ENSDART00000147678
|
CR769768.1
|
|
| chr3_+_30257582 | 0.28 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr7_+_41314862 | 0.28 |
ENSDART00000185198
|
zgc:165532
|
zgc:165532 |
| chr23_+_44634187 | 0.27 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr3_+_34159192 | 0.27 |
ENSDART00000151119
|
carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr6_+_60036767 | 0.26 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr8_+_21159122 | 0.25 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr6_+_9872261 | 0.25 |
ENSDART00000045070
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr3_+_19336286 | 0.25 |
ENSDART00000111528
|
kri1
|
KRI1 homolog |
| chr8_-_11146687 | 0.24 |
ENSDART00000189626
|
asb13b
|
ankyrin repeat and SOCS box containing 13b |
| chr1_-_44314 | 0.24 |
ENSDART00000160050
|
tmem39a
|
transmembrane protein 39A |
| chr23_-_2448234 | 0.24 |
ENSDART00000082097
|
LO018513.1
|
|
| chr2_+_6067371 | 0.23 |
ENSDART00000053868
ENSDART00000145244 |
aldh9a1b
|
aldehyde dehydrogenase 9 family, member A1b |
| chr8_+_8699085 | 0.21 |
ENSDART00000021209
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr7_-_29231686 | 0.21 |
ENSDART00000173780
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr15_-_1844048 | 0.21 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr12_-_4176329 | 0.21 |
ENSDART00000092746
|
si:dkey-32n7.7
|
si:dkey-32n7.7 |
| chr25_+_12012 | 0.20 |
ENSDART00000170348
|
cntn1a
|
contactin 1a |
| chr20_-_25877793 | 0.19 |
ENSDART00000177333
ENSDART00000184619 |
exoc1
|
exocyst complex component 1 |
| chr11_+_36243774 | 0.19 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr2_+_21486529 | 0.18 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr20_-_26936887 | 0.17 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr22_+_17399124 | 0.15 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr23_+_4583747 | 0.14 |
ENSDART00000160350
|
LO017700.1
|
|
| chr17_-_46850299 | 0.13 |
ENSDART00000154430
|
pimr23
|
Pim proto-oncogene, serine/threonine kinase, related 23 |
| chr2_-_56649883 | 0.13 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr1_+_58353661 | 0.12 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr17_+_52524452 | 0.11 |
ENSDART00000016614
|
dlst
|
dihydrolipoamide S-succinyltransferase |
| chr8_+_2530065 | 0.11 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr13_-_31878043 | 0.09 |
ENSDART00000187720
|
syt14a
|
synaptotagmin XIVa |
| chr18_+_43183749 | 0.09 |
ENSDART00000151166
|
nectin1b
|
nectin cell adhesion molecule 1b |
| chr3_+_18424244 | 0.09 |
ENSDART00000127796
|
gaa
|
glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) |
| chr9_+_34433053 | 0.08 |
ENSDART00000137416
|
si:ch211-269e2.1
|
si:ch211-269e2.1 |
| chr2_+_43750899 | 0.07 |
ENSDART00000135836
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr20_+_36806398 | 0.06 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr14_+_22293787 | 0.05 |
ENSDART00000157967
|
atp10b
|
ATPase phospholipid transporting 10B |
| chr19_-_3193443 | 0.04 |
ENSDART00000179855
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr4_-_20243021 | 0.03 |
ENSDART00000048023
|
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr12_+_19138452 | 0.03 |
ENSDART00000141346
ENSDART00000066397 |
phf5a
|
PHD finger protein 5A |
| chr1_+_58221372 | 0.02 |
ENSDART00000138467
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr17_-_46933567 | 0.02 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr19_+_20274944 | 0.01 |
ENSDART00000151237
|
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr8_+_8893535 | 0.01 |
ENSDART00000139150
|
otud5a
|
OTU deubiquitinase 5a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2c1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 1.1 | 3.3 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.6 | 2.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.6 | 2.3 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.5 | 1.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.5 | 2.5 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.5 | 1.4 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.4 | 1.8 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.4 | 3.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 1.7 | GO:0010259 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 1.7 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.3 | 2.8 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.3 | 1.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.2 | 2.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 0.5 | GO:0042671 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.2 | 0.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 1.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 1.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.2 | 0.7 | GO:0051610 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.2 | 0.6 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.2 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 1.9 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.7 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 2.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.4 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 1.1 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.4 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.1 | 0.4 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 1.2 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 1.9 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.5 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 0.9 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 1.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.5 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.1 | 0.8 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 3.9 | GO:0048599 | oocyte development(GO:0048599) |
| 0.1 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.7 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 0.7 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.1 | 0.5 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.3 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 0.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 1.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 3.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.7 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 1.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 1.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.9 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0044406 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.2 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 1.0 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 1.7 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.7 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.6 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.9 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.7 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 1.1 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.5 | 2.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.1 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.7 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 4.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.9 | 2.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.8 | 3.8 | GO:0009374 | biotin binding(GO:0009374) |
| 0.6 | 2.4 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.4 | 1.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.4 | 1.3 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 1.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.3 | 2.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.3 | 1.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.3 | 1.0 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.2 | 0.7 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 3.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 1.7 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 2.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 1.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 1.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.7 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 2.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.3 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.2 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.1 | 0.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.9 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 4.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.3 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.1 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.7 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.2 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 2.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 4.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 2.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.9 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 2.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 1.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 3.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 2.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 2.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.2 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |