Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
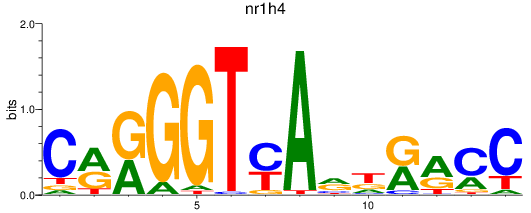
Results for nr1h4
Z-value: 0.49
Transcription factors associated with nr1h4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr1h4
|
ENSDARG00000057741 | nuclear receptor subfamily 1, group H, member 4 |
|
nr1h4
|
ENSDARG00000113600 | nuclear receptor subfamily 1, group H, member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr1h4 | dr11_v1_chr18_+_15644559_15644559 | -0.37 | 3.3e-01 | Click! |
Activity profile of nr1h4 motif
Sorted Z-values of nr1h4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_20693724 | 1.08 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr7_+_39399747 | 0.97 |
ENSDART00000147037
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr5_+_4366431 | 0.86 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr13_-_7766758 | 0.83 |
ENSDART00000171831
|
h2afy2
|
H2A histone family, member Y2 |
| chr9_+_48007081 | 0.73 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr8_+_1769475 | 0.72 |
ENSDART00000079073
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr19_+_32166702 | 0.71 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr2_-_42234484 | 0.70 |
ENSDART00000132617
ENSDART00000136690 ENSDART00000141358 |
apom
|
apolipoprotein M |
| chr12_-_30841679 | 0.68 |
ENSDART00000105594
|
crygmx
|
crystallin, gamma MX |
| chr1_-_44940830 | 0.67 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr25_+_23591990 | 0.62 |
ENSDART00000151938
ENSDART00000089199 |
cpt1ab
|
carnitine palmitoyltransferase 1Ab (liver) |
| chr21_-_5879897 | 0.61 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr23_+_35684756 | 0.59 |
ENSDART00000132320
|
pmelb
|
premelanosome protein b |
| chr8_+_10305400 | 0.56 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr3_-_19899914 | 0.55 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr11_-_6974022 | 0.54 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr4_+_1530287 | 0.50 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr10_-_27197044 | 0.49 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr1_+_127250 | 0.49 |
ENSDART00000003463
|
f7i
|
coagulation factor VIIi |
| chr10_+_22012389 | 0.46 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr6_-_40312091 | 0.43 |
ENSDART00000154878
|
col7a1
|
collagen, type VII, alpha 1 |
| chr7_-_52531252 | 0.43 |
ENSDART00000174369
|
tcf12
|
transcription factor 12 |
| chr11_+_11200550 | 0.43 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr16_-_13730152 | 0.42 |
ENSDART00000138772
|
ttyh1
|
tweety family member 1 |
| chr14_+_34514336 | 0.42 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr16_+_5184402 | 0.41 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr5_+_1219187 | 0.41 |
ENSDART00000129490
|
gb:bc139872
|
expressed sequence BC139872 |
| chr16_-_7228276 | 0.41 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr16_-_20312146 | 0.40 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr11_+_21910752 | 0.40 |
ENSDART00000114288
|
foxp4
|
forkhead box P4 |
| chr11_-_22372072 | 0.39 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr22_+_10645088 | 0.38 |
ENSDART00000142265
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr5_+_69716458 | 0.38 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr13_-_37108310 | 0.37 |
ENSDART00000163845
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr1_+_44941031 | 0.37 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr24_+_28448387 | 0.36 |
ENSDART00000154077
ENSDART00000018037 |
arhgap29a
|
Rho GTPase activating protein 29a |
| chr20_-_19864131 | 0.36 |
ENSDART00000057819
|
ptk2bb
|
protein tyrosine kinase 2 beta, b |
| chr25_-_24046870 | 0.35 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr8_-_13013123 | 0.35 |
ENSDART00000147802
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr16_+_53203370 | 0.34 |
ENSDART00000154669
|
si:ch211-269k10.2
|
si:ch211-269k10.2 |
| chr2_-_6051836 | 0.33 |
ENSDART00000092479
|
si:ch211-284b7.3
|
si:ch211-284b7.3 |
| chr4_-_16545085 | 0.33 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr9_-_1986014 | 0.33 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr4_-_5597167 | 0.33 |
ENSDART00000132431
|
vegfab
|
vascular endothelial growth factor Ab |
| chr18_+_22302635 | 0.33 |
ENSDART00000141051
|
carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr3_-_4303262 | 0.32 |
ENSDART00000112819
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr16_-_50181206 | 0.31 |
ENSDART00000148478
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr19_-_18855513 | 0.30 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr15_+_6652396 | 0.30 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr1_-_17650223 | 0.29 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr5_+_65946222 | 0.29 |
ENSDART00000190969
ENSDART00000161578 |
mymk
|
myomaker, myoblast fusion factor |
| chr1_-_21344478 | 0.28 |
ENSDART00000077805
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr11_+_691734 | 0.27 |
ENSDART00000191463
|
timp4.1
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 1 |
| chr23_+_43954809 | 0.27 |
ENSDART00000164080
|
corin
|
corin, serine peptidase |
| chr22_+_8313513 | 0.27 |
ENSDART00000181169
ENSDART00000103911 |
CABZ01077217.1
|
|
| chr14_-_3174570 | 0.27 |
ENSDART00000163585
|
csf1ra
|
colony stimulating factor 1 receptor, a |
| chr12_+_34119439 | 0.26 |
ENSDART00000032821
|
cyth1b
|
cytohesin 1b |
| chr21_+_44112914 | 0.26 |
ENSDART00000062836
|
fgf1b
|
fibroblast growth factor 1b |
| chr24_-_21689146 | 0.25 |
ENSDART00000105917
|
urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5-) decarboxylase |
| chr5_+_25072894 | 0.25 |
ENSDART00000012268
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr24_+_28528000 | 0.25 |
ENSDART00000155924
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr17_+_996509 | 0.24 |
ENSDART00000158830
|
cyp1c2
|
cytochrome P450, family 1, subfamily C, polypeptide 2 |
| chr19_-_2861444 | 0.24 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr6_-_609880 | 0.24 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr15_-_18091157 | 0.24 |
ENSDART00000153848
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr19_-_18855717 | 0.23 |
ENSDART00000158192
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr5_+_1079423 | 0.23 |
ENSDART00000172231
|
si:zfos-128g4.2
|
si:zfos-128g4.2 |
| chr7_+_34688527 | 0.23 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr20_-_4738101 | 0.22 |
ENSDART00000050201
ENSDART00000152559 ENSDART00000053858 ENSDART00000125620 |
papola
|
poly(A) polymerase alpha |
| chr18_+_39067575 | 0.22 |
ENSDART00000077724
|
gnb5b
|
guanine nucleotide binding protein (G protein), beta 5b |
| chr11_+_5842632 | 0.21 |
ENSDART00000111374
ENSDART00000158599 |
ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, (NADH-coenzyme Q reductase) |
| chr12_+_695619 | 0.21 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr15_-_19705707 | 0.20 |
ENSDART00000047643
|
sytl2b
|
synaptotagmin-like 2b |
| chr25_+_32473277 | 0.20 |
ENSDART00000146451
|
sqor
|
sulfide quinone oxidoreductase |
| chr23_-_45504991 | 0.20 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr22_-_11136625 | 0.19 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr13_+_29238850 | 0.19 |
ENSDART00000026000
|
myofl
|
myoferlin like |
| chr25_+_245018 | 0.19 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr14_-_34771371 | 0.19 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr16_-_4769877 | 0.19 |
ENSDART00000149421
ENSDART00000054078 |
rpa2
|
replication protein A2 |
| chr23_+_42810055 | 0.19 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr23_+_7471072 | 0.18 |
ENSDART00000135551
|
si:ch211-200e2.1
|
si:ch211-200e2.1 |
| chr17_+_20173882 | 0.18 |
ENSDART00000155379
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr21_+_27558381 | 0.17 |
ENSDART00000024287
ENSDART00000144869 |
zgc:165604
|
zgc:165604 |
| chr5_-_25072607 | 0.17 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr3_-_58582663 | 0.17 |
ENSDART00000180055
|
CABZ01038521.1
|
|
| chr25_-_16969922 | 0.17 |
ENSDART00000111158
|
tigara
|
tp53-induced glycolysis and apoptosis regulator a |
| chr16_+_14684916 | 0.16 |
ENSDART00000138611
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr9_-_28616436 | 0.16 |
ENSDART00000136985
|
si:ch73-7i4.2
|
si:ch73-7i4.2 |
| chr25_+_12494079 | 0.16 |
ENSDART00000163508
|
map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr19_-_1929310 | 0.15 |
ENSDART00000144113
|
si:ch211-149a19.3
|
si:ch211-149a19.3 |
| chr20_-_37522569 | 0.15 |
ENSDART00000177011
ENSDART00000058502 |
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr21_-_12036134 | 0.15 |
ENSDART00000031658
|
tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr22_+_21252790 | 0.15 |
ENSDART00000079046
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr3_-_18030938 | 0.15 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr21_+_29179887 | 0.15 |
ENSDART00000161941
|
si:ch211-57b15.1
|
si:ch211-57b15.1 |
| chr20_+_54685143 | 0.14 |
ENSDART00000162535
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr6_+_9677420 | 0.14 |
ENSDART00000064995
|
sumo1
|
small ubiquitin-like modifier 1 |
| chr7_-_41338923 | 0.14 |
ENSDART00000099138
|
ncf2
|
neutrophil cytosolic factor 2 |
| chr7_+_11197940 | 0.14 |
ENSDART00000081346
|
cemip
|
cell migration inducing protein, hyaluronan binding |
| chr4_-_18455480 | 0.14 |
ENSDART00000150221
|
sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr19_-_20199167 | 0.14 |
ENSDART00000155527
|
si:ch211-155k24.9
|
si:ch211-155k24.9 |
| chr5_+_38886499 | 0.14 |
ENSDART00000076845
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr23_-_22598002 | 0.14 |
ENSDART00000146966
|
si:ch211-28e16.5
|
si:ch211-28e16.5 |
| chr15_-_14552101 | 0.13 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr21_-_20733615 | 0.13 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr7_-_69983462 | 0.13 |
ENSDART00000123380
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr24_+_25467465 | 0.13 |
ENSDART00000189933
|
smpx
|
small muscle protein, X-linked |
| chr6_-_1514767 | 0.13 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr3_+_4346854 | 0.13 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr20_+_7243879 | 0.12 |
ENSDART00000122298
|
bsnd
|
barttin CLCNK-type chloride channel accessory beta subunit |
| chr25_+_5012791 | 0.12 |
ENSDART00000156970
|
si:ch73-265h17.5
|
si:ch73-265h17.5 |
| chr10_+_20070178 | 0.12 |
ENSDART00000027612
ENSDART00000145264 ENSDART00000172713 |
xpo7
|
exportin 7 |
| chr2_-_37862380 | 0.12 |
ENSDART00000186005
|
si:ch211-284o19.8
|
si:ch211-284o19.8 |
| chr9_+_2499627 | 0.12 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr3_+_12835370 | 0.12 |
ENSDART00000166089
|
si:ch211-8c17.3
|
si:ch211-8c17.3 |
| chr7_-_8014186 | 0.12 |
ENSDART00000190012
|
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr13_+_35746440 | 0.12 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr23_-_21515182 | 0.12 |
ENSDART00000142000
|
rnf207b
|
ring finger protein 207b |
| chr10_-_20696518 | 0.11 |
ENSDART00000064581
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr16_+_49091322 | 0.11 |
ENSDART00000186400
|
plcl2
|
phospholipase C like 2 |
| chr13_-_337318 | 0.11 |
ENSDART00000166175
|
zgc:171534
|
zgc:171534 |
| chr3_-_49110710 | 0.11 |
ENSDART00000160404
|
trim35-12
|
tripartite motif containing 35-12 |
| chr3_-_34027178 | 0.11 |
ENSDART00000170201
ENSDART00000151408 |
ighv1-4
ighv14-1
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 14-1 |
| chr22_-_11137268 | 0.10 |
ENSDART00000178882
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr22_+_25590391 | 0.10 |
ENSDART00000178133
|
aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr17_+_14711765 | 0.09 |
ENSDART00000012889
|
cx28.6
|
connexin 28.6 |
| chr15_-_17619306 | 0.09 |
ENSDART00000184011
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr13_-_37200120 | 0.09 |
ENSDART00000135001
|
si:dkeyp-77c8.3
|
si:dkeyp-77c8.3 |
| chr5_-_50748081 | 0.09 |
ENSDART00000113985
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr5_-_16218777 | 0.09 |
ENSDART00000141698
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr1_-_22512063 | 0.09 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr15_-_2857961 | 0.09 |
ENSDART00000033263
|
ankrd49
|
ankyrin repeat domain 49 |
| chr22_-_36926342 | 0.08 |
ENSDART00000151804
|
si:dkey-37m8.11
|
si:dkey-37m8.11 |
| chr4_-_11165611 | 0.08 |
ENSDART00000059489
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr3_+_32142382 | 0.08 |
ENSDART00000133035
|
syt5a
|
synaptotagmin Va |
| chr5_+_54400971 | 0.08 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr7_+_52811291 | 0.08 |
ENSDART00000192900
|
CT030160.1
|
|
| chr12_-_25201576 | 0.07 |
ENSDART00000077188
|
cox7a3
|
cytochrome c oxidase subunit VIIa polypeptide 3 |
| chr11_-_41786522 | 0.07 |
ENSDART00000191066
|
CR847970.1
|
|
| chr22_+_7486867 | 0.07 |
ENSDART00000034586
|
CELA1 (1 of many)
|
zgc:112302 |
| chr3_+_31953145 | 0.07 |
ENSDART00000148861
|
kcnc3a
|
potassium voltage-gated channel, Shaw-related subfamily, member 3a |
| chr20_+_9828769 | 0.07 |
ENSDART00000160480
ENSDART00000053844 ENSDART00000080691 |
stxbp6l
|
syntaxin binding protein 6 (amisyn), like |
| chr16_-_50157187 | 0.07 |
ENSDART00000155162
|
nek10
|
NIMA-related kinase 10 |
| chr9_+_38158570 | 0.06 |
ENSDART00000059549
ENSDART00000133060 |
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr25_+_32473433 | 0.06 |
ENSDART00000152326
|
sqor
|
sulfide quinone oxidoreductase |
| chr6_+_9398074 | 0.06 |
ENSDART00000151772
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr10_+_3507861 | 0.06 |
ENSDART00000092684
|
rph3aa
|
rabphilin 3A homolog (mouse), a |
| chr4_-_11165961 | 0.06 |
ENSDART00000193393
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr17_-_465285 | 0.05 |
ENSDART00000168718
|
chrm5a
|
cholinergic receptor, muscarinic 5a |
| chr7_+_3597413 | 0.05 |
ENSDART00000064281
|
si:dkey-192d15.2
|
si:dkey-192d15.2 |
| chr6_+_52869892 | 0.05 |
ENSDART00000146143
|
si:dkeyp-3f10.16
|
si:dkeyp-3f10.16 |
| chr18_-_41754020 | 0.05 |
ENSDART00000077898
|
grk7b
|
G protein-coupled receptor kinase 7b |
| chr9_-_16877456 | 0.05 |
ENSDART00000161105
ENSDART00000160869 |
fbxl3a
|
F-box and leucine-rich repeat protein 3a |
| chr9_+_15890558 | 0.05 |
ENSDART00000144032
|
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr19_+_2685779 | 0.05 |
ENSDART00000160533
ENSDART00000097531 |
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr18_+_3338228 | 0.05 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr17_-_25395395 | 0.05 |
ENSDART00000170233
|
fam167b
|
family with sequence similarity 167, member B |
| chr15_-_37752236 | 0.05 |
ENSDART00000154263
|
si:dkey-42l23.5
|
si:dkey-42l23.5 |
| chr8_-_36469117 | 0.05 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr15_-_462783 | 0.05 |
ENSDART00000161049
|
CABZ01033205.1
|
Danio rerio uncharacterized LOC100002960 (LOC100002960), mRNA. |
| chr11_+_12175162 | 0.05 |
ENSDART00000125446
|
si:ch211-156l18.7
|
si:ch211-156l18.7 |
| chr1_-_44704261 | 0.04 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr4_+_60492313 | 0.04 |
ENSDART00000191043
ENSDART00000191631 |
si:dkey-211i20.2
|
si:dkey-211i20.2 |
| chr19_-_23369282 | 0.04 |
ENSDART00000147824
|
pimr90
|
Pim proto-oncogene, serine/threonine kinase, related 90 |
| chr13_-_30713236 | 0.04 |
ENSDART00000112372
ENSDART00000142221 |
tmem72
|
transmembrane protein 72 |
| chr4_-_38281909 | 0.04 |
ENSDART00000162448
|
si:ch211-229l10.9
|
si:ch211-229l10.9 |
| chr5_+_16496273 | 0.04 |
ENSDART00000168969
ENSDART00000147442 |
htr7c
|
5-hydroxytryptamine (serotonin) receptor 7c |
| chr7_+_29153018 | 0.04 |
ENSDART00000138065
|
tmppe
|
transmembrane protein with metallophosphoesterase domain |
| chr15_+_45563656 | 0.03 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr14_-_2318590 | 0.03 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr7_-_1918971 | 0.03 |
ENSDART00000181759
|
CABZ01040626.1
|
|
| chrM_-_15232 | 0.03 |
ENSDART00000093623
|
mt-nd6
|
NADH dehydrogenase 6, mitochondrial |
| chr24_-_25184553 | 0.03 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr11_-_5563498 | 0.03 |
ENSDART00000160835
|
pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr7_+_67494107 | 0.03 |
ENSDART00000185653
|
cpne7
|
copine VII |
| chr6_+_41956355 | 0.03 |
ENSDART00000176856
|
oxtr
|
oxytocin receptor |
| chr1_+_58353661 | 0.03 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr14_-_34276574 | 0.03 |
ENSDART00000021437
|
gria1a
|
glutamate receptor, ionotropic, AMPA 1a |
| chr18_+_31056645 | 0.02 |
ENSDART00000159316
|
mvda
|
mevalonate (diphospho) decarboxylase a |
| chr7_-_16237861 | 0.02 |
ENSDART00000173647
|
btr07
|
bloodthirsty-related gene family, member 7 |
| chr15_-_34322915 | 0.02 |
ENSDART00000190543
ENSDART00000019651 ENSDART00000193601 |
dgkb
|
diacylglycerol kinase, beta |
| chr14_+_904850 | 0.02 |
ENSDART00000161847
|
si:ch73-208h1.1
|
si:ch73-208h1.1 |
| chr6_+_15145123 | 0.02 |
ENSDART00000063648
|
nck2b
|
NCK adaptor protein 2b |
| chr1_+_56768263 | 0.02 |
ENSDART00000158530
|
CABZ01049362.1
|
|
| chr21_-_22664554 | 0.02 |
ENSDART00000161438
|
gig2k
|
grass carp reovirus (GCRV)-induced gene 2k |
| chr2_+_371759 | 0.02 |
ENSDART00000153788
|
si:dkey-33c14.7
|
si:dkey-33c14.7 |
| chr22_+_34663328 | 0.02 |
ENSDART00000183912
|
si:ch73-304f21.1
|
si:ch73-304f21.1 |
| chr15_+_37992275 | 0.02 |
ENSDART00000154928
|
si:dkey-238d18.7
|
si:dkey-238d18.7 |
| chr25_-_255971 | 0.02 |
ENSDART00000162006
|
CU855936.1
|
|
| chr2_-_52020049 | 0.02 |
ENSDART00000128198
ENSDART00000135331 |
scn12aa
|
sodium channel, voltage gated, type XII, alpha a |
| chr1_+_58323980 | 0.01 |
ENSDART00000145743
|
si:dkey-222h21.3
|
si:dkey-222h21.3 |
| chr2_+_42112503 | 0.01 |
ENSDART00000136022
|
crha
|
corticotropin releasing hormone a |
| chr16_-_11789108 | 0.01 |
ENSDART00000060142
|
tlr21
|
toll-like receptor 21 |
| chr16_-_17175731 | 0.01 |
ENSDART00000183057
|
opn9
|
opsin 9 |
| chr13_-_11699530 | 0.01 |
ENSDART00000192161
|
slc39a8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr15_-_31000275 | 0.01 |
ENSDART00000132493
ENSDART00000100183 |
lgals9l6
lgals9l5
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 6 lectin, galactoside-binding, soluble, 9 (galectin 9)-like 5 |
| chr7_+_37359004 | 0.01 |
ENSDART00000192134
ENSDART00000189239 |
sall1a
|
spalt-like transcription factor 1a |
| chr3_-_4253214 | 0.01 |
ENSDART00000155807
|
si:dkey-175d9.2
|
si:dkey-175d9.2 |
| chr12_-_4370585 | 0.00 |
ENSDART00000129502
|
CA4 (1 of many)
|
si:ch211-173d10.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr1h4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.4 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.3 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.3 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.1 | 0.3 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 0.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.3 | GO:0010759 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) regulation of odontogenesis(GO:0042481) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.1 | 0.6 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.2 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.1 | 0.2 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.3 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.6 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.4 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.0 | 0.3 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.6 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.4 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.3 | GO:0030324 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.3 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.2 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.1 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |