Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
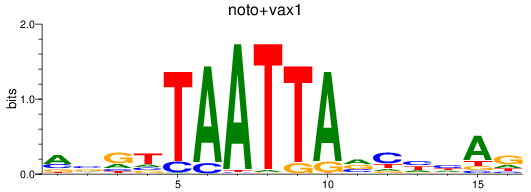
Results for noto+vax1
Z-value: 0.46
Transcription factors associated with noto+vax1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
noto
|
ENSDARG00000021201 | notochord homeobox |
|
vax1
|
ENSDARG00000021916 | ventral anterior homeobox 1 |
|
vax1
|
ENSDARG00000114230 | ventral anterior homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| noto | dr11_v1_chr13_+_14976108_14976108 | -0.68 | 4.3e-02 | Click! |
| vax1 | dr11_v1_chr17_-_21441464_21441464 | 0.07 | 8.6e-01 | Click! |
Activity profile of noto+vax1 motif
Sorted Z-values of noto+vax1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_47446158 | 0.65 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr5_+_20257225 | 0.64 |
ENSDART00000127919
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr13_-_35808904 | 0.63 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr19_-_18313303 | 0.61 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr2_+_50608099 | 0.59 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr19_+_41464870 | 0.47 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr14_+_51056605 | 0.46 |
ENSDART00000159639
|
CABZ01078593.1
|
|
| chr5_-_15283509 | 0.46 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr9_-_14683574 | 0.45 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr8_-_22274222 | 0.45 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr22_-_21897203 | 0.43 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr2_-_38284648 | 0.42 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr20_-_18382708 | 0.39 |
ENSDART00000170864
ENSDART00000166762 ENSDART00000191333 |
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr6_+_32382743 | 0.37 |
ENSDART00000190009
|
dock7
|
dedicator of cytokinesis 7 |
| chr19_+_32401278 | 0.35 |
ENSDART00000184353
|
atxn1a
|
ataxin 1a |
| chr16_-_22544047 | 0.34 |
ENSDART00000131657
|
cgna
|
cingulin a |
| chr16_-_7828838 | 0.31 |
ENSDART00000191434
ENSDART00000108653 |
tcaim
|
T cell activation inhibitor, mitochondrial |
| chr25_+_2361721 | 0.30 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr11_-_13107106 | 0.29 |
ENSDART00000184477
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr6_+_40354424 | 0.29 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr18_+_5137241 | 0.28 |
ENSDART00000159601
|
serpini1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr20_-_1635922 | 0.28 |
ENSDART00000181502
|
CR846082.1
|
|
| chr18_-_43884044 | 0.27 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr19_-_20430892 | 0.27 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr15_+_31701716 | 0.27 |
ENSDART00000113370
|
b3glcta
|
beta 3-glucosyltransferase a |
| chr7_-_12464412 | 0.26 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr17_-_3815693 | 0.26 |
ENSDART00000161207
|
PLCB4
|
phospholipase C beta 4 |
| chr10_+_28160265 | 0.25 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr10_-_2524917 | 0.24 |
ENSDART00000188642
|
CU856539.1
|
|
| chr19_+_2631565 | 0.24 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr6_-_57539141 | 0.24 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr12_+_48803098 | 0.23 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr21_+_8427059 | 0.23 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr12_+_34891529 | 0.23 |
ENSDART00000015643
|
tbcc
|
tubulin folding cofactor C |
| chr8_-_25034411 | 0.22 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr8_-_52562672 | 0.21 |
ENSDART00000159333
ENSDART00000159974 |
si:ch73-199g24.2
|
si:ch73-199g24.2 |
| chr4_+_9011448 | 0.21 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr8_-_39822917 | 0.20 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr6_-_19271210 | 0.20 |
ENSDART00000163628
ENSDART00000159124 |
zgc:174863
|
zgc:174863 |
| chr2_-_43123949 | 0.20 |
ENSDART00000075347
ENSDART00000132346 |
lrrc6
|
leucine rich repeat containing 6 |
| chr11_+_11303458 | 0.19 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr2_-_39558643 | 0.19 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr2_-_2957970 | 0.19 |
ENSDART00000162505
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr4_+_21129752 | 0.18 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr15_+_44366556 | 0.17 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr13_-_24257631 | 0.15 |
ENSDART00000146524
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr17_+_21902058 | 0.15 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr1_+_513986 | 0.15 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr2_-_26720854 | 0.14 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr6_-_30954268 | 0.13 |
ENSDART00000154523
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr5_+_41477954 | 0.11 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr12_+_22580579 | 0.11 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr9_-_54248182 | 0.11 |
ENSDART00000129540
|
larsa
|
leucyl-tRNA synthetase a |
| chr10_-_33621739 | 0.11 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr18_-_2727764 | 0.11 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr6_-_40922971 | 0.10 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr16_+_43765993 | 0.10 |
ENSDART00000149154
|
si:dkey-122a22.2
|
si:dkey-122a22.2 |
| chr14_+_22591624 | 0.09 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr3_-_32902138 | 0.08 |
ENSDART00000144026
ENSDART00000083874 ENSDART00000145443 ENSDART00000148239 ENSDART00000134917 |
kat7a
|
K(lysine) acetyltransferase 7a |
| chr10_+_16036246 | 0.08 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr20_+_29209767 | 0.08 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr14_+_30762131 | 0.08 |
ENSDART00000145039
|
si:ch211-145o7.3
|
si:ch211-145o7.3 |
| chr19_+_9295244 | 0.07 |
ENSDART00000132255
ENSDART00000144299 |
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr20_-_37813863 | 0.07 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr6_+_3280939 | 0.07 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr23_-_1348933 | 0.07 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr7_-_46777876 | 0.06 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr5_+_41477526 | 0.06 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr16_-_13818061 | 0.06 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr22_-_2959005 | 0.05 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr17_+_16046314 | 0.05 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr18_+_9362455 | 0.05 |
ENSDART00000187025
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr1_-_31534089 | 0.05 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr10_-_26744131 | 0.05 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr13_+_15190677 | 0.05 |
ENSDART00000142240
ENSDART00000129045 |
mavs
|
mitochondrial antiviral signaling protein |
| chr7_+_65261576 | 0.04 |
ENSDART00000169566
|
bco1
|
beta-carotene oxygenase 1 |
| chr18_+_17827149 | 0.04 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr24_+_19415124 | 0.03 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr6_-_40581376 | 0.03 |
ENSDART00000185412
|
tspo
|
translocator protein |
| chr2_-_21819421 | 0.03 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr15_+_31899312 | 0.03 |
ENSDART00000155315
|
frya
|
furry homolog a (Drosophila) |
| chr1_-_7970653 | 0.03 |
ENSDART00000188909
|
si:dkey-79f11.7
|
si:dkey-79f11.7 |
| chr25_+_36338289 | 0.03 |
ENSDART00000187881
|
zgc:165555
|
zgc:165555 |
| chr21_-_42876565 | 0.02 |
ENSDART00000126480
|
zmp:0000001268
|
zmp:0000001268 |
| chr16_+_45425181 | 0.02 |
ENSDART00000168591
|
phf1
|
PHD finger protein 1 |
| chr8_+_25034544 | 0.01 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr1_-_51719110 | 0.01 |
ENSDART00000190574
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr3_-_27601812 | 0.01 |
ENSDART00000077734
|
zgc:66160
|
zgc:66160 |
| chr13_+_8677166 | 0.01 |
ENSDART00000181016
ENSDART00000135028 |
prop1
|
PROP paired-like homeobox 1 |
| chr14_-_36320506 | 0.01 |
ENSDART00000074639
|
egf
|
epidermal growth factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of noto+vax1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.2 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.3 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.4 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.5 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |