Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
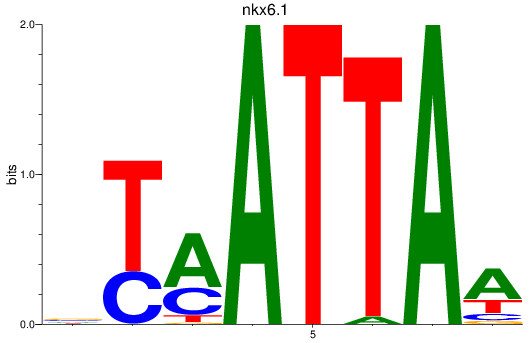
Results for nkx6.1
Z-value: 0.68
Transcription factors associated with nkx6.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx6.1
|
ENSDARG00000022569 | NK6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx6.1 | dr11_v1_chr21_+_19070921_19070921 | 0.01 | 9.8e-01 | Click! |
Activity profile of nkx6.1 motif
Sorted Z-values of nkx6.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_30244356 | 1.21 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr1_-_43905252 | 0.79 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr7_+_14291323 | 0.58 |
ENSDART00000053521
|
rhcga
|
Rh family, C glycoprotein a |
| chr25_+_29160102 | 0.56 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr3_-_50443607 | 0.47 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr16_+_46111849 | 0.46 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr5_-_30620625 | 0.44 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr20_-_40755614 | 0.41 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr17_+_15433671 | 0.39 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr2_-_27329214 | 0.39 |
ENSDART00000145835
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr17_+_15433518 | 0.39 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr5_-_23362602 | 0.37 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr12_-_35054354 | 0.35 |
ENSDART00000075351
|
zgc:112285
|
zgc:112285 |
| chr16_+_23431189 | 0.35 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr2_+_37227011 | 0.35 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr7_-_28148310 | 0.33 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr18_+_46773198 | 0.33 |
ENSDART00000174647
|
CABZ01041495.1
|
|
| chr5_-_44829719 | 0.32 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr19_+_10339538 | 0.31 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr14_+_49135264 | 0.30 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr10_+_22381802 | 0.29 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr12_+_24342303 | 0.28 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr13_+_29462249 | 0.28 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr23_+_40460333 | 0.27 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr16_-_45178430 | 0.26 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr3_+_23092762 | 0.25 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr1_-_43915423 | 0.25 |
ENSDART00000181915
ENSDART00000113673 |
scpp5
|
secretory calcium-binding phosphoprotein 5 |
| chr20_+_18551657 | 0.25 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr1_-_22512063 | 0.24 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr16_+_23913943 | 0.23 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr12_+_5081759 | 0.23 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr5_-_38155005 | 0.23 |
ENSDART00000097770
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr13_+_23988442 | 0.23 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr22_+_28320792 | 0.22 |
ENSDART00000160562
ENSDART00000164138 |
impg2b
impg2b
|
interphotoreceptor matrix proteoglycan 2b interphotoreceptor matrix proteoglycan 2b |
| chr14_+_50770537 | 0.22 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr19_+_32166702 | 0.22 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr21_+_6780340 | 0.22 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr13_+_38430466 | 0.22 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr25_+_3058924 | 0.22 |
ENSDART00000029580
|
fth1b
|
ferritin, heavy polypeptide 1b |
| chr7_-_24699985 | 0.22 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr15_+_22311803 | 0.21 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr8_-_34052019 | 0.21 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr5_-_43071058 | 0.21 |
ENSDART00000165546
|
si:dkey-245n4.2
|
si:dkey-245n4.2 |
| chr3_-_41791178 | 0.21 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr17_-_12389259 | 0.21 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr3_-_28258462 | 0.20 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr22_-_13042992 | 0.20 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr14_-_2933185 | 0.20 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr21_-_22827548 | 0.20 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr8_-_25120231 | 0.20 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr13_-_30700460 | 0.19 |
ENSDART00000139073
|
rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr7_-_28147838 | 0.19 |
ENSDART00000158921
|
lmo1
|
LIM domain only 1 |
| chr9_+_34641237 | 0.19 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr21_-_42100471 | 0.19 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr10_+_25726694 | 0.19 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr20_-_27711970 | 0.19 |
ENSDART00000139637
|
zbtb25
|
zinc finger and BTB domain containing 25 |
| chr3_+_28953274 | 0.19 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr5_+_63668735 | 0.18 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr6_-_37469775 | 0.18 |
ENSDART00000156546
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr17_-_16965809 | 0.18 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr1_-_50859053 | 0.18 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr6_-_35779348 | 0.18 |
ENSDART00000191159
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr4_+_21129752 | 0.18 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr12_+_18681477 | 0.18 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr14_-_4556896 | 0.18 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr23_-_10175898 | 0.18 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr22_+_12770877 | 0.18 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr22_+_20427170 | 0.17 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr2_+_6963296 | 0.17 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr17_-_19019635 | 0.17 |
ENSDART00000126666
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr16_+_5774977 | 0.17 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr1_-_36152131 | 0.17 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr10_+_29698467 | 0.17 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr15_+_21276735 | 0.17 |
ENSDART00000111213
|
ubash3bb
|
ubiquitin associated and SH3 domain containing Bb |
| chr10_+_37137464 | 0.17 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr1_+_52929185 | 0.16 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr8_-_25329967 | 0.16 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr1_+_25801648 | 0.16 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr17_+_29276544 | 0.16 |
ENSDART00000049399
|
ankrd9
|
ankyrin repeat domain 9 |
| chr2_+_50608099 | 0.16 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr10_+_32683089 | 0.16 |
ENSDART00000063551
|
ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr13_+_38521152 | 0.16 |
ENSDART00000145292
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr11_+_30057762 | 0.16 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr23_+_28731379 | 0.16 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr22_-_24738188 | 0.15 |
ENSDART00000050238
|
vtg1
|
vitellogenin 1 |
| chr12_+_31713239 | 0.15 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr10_-_8060573 | 0.15 |
ENSDART00000147104
ENSDART00000099030 |
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr10_-_26512993 | 0.15 |
ENSDART00000188549
ENSDART00000193316 |
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr11_+_30000814 | 0.15 |
ENSDART00000191011
ENSDART00000189770 |
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr8_-_42238543 | 0.15 |
ENSDART00000062697
|
gfra2a
|
GDNF family receptor alpha 2a |
| chr5_-_28109416 | 0.15 |
ENSDART00000042002
|
si:ch211-48m9.1
|
si:ch211-48m9.1 |
| chr15_+_36457888 | 0.15 |
ENSDART00000155100
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr4_+_12615836 | 0.15 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr1_+_33969015 | 0.15 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr6_-_10320676 | 0.15 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr15_-_6247775 | 0.14 |
ENSDART00000148350
|
dscamb
|
Down syndrome cell adhesion molecule b |
| chr1_+_12766351 | 0.14 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr14_+_45406299 | 0.14 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr23_+_21638258 | 0.14 |
ENSDART00000104188
|
igsf21b
|
immunoglobin superfamily, member 21b |
| chr18_-_48550426 | 0.14 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr21_+_28445052 | 0.14 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr12_-_19007834 | 0.14 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr22_-_24791505 | 0.14 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr18_-_1185772 | 0.14 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr20_+_4060839 | 0.14 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr18_+_15644559 | 0.14 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr14_+_14662116 | 0.14 |
ENSDART00000161693
|
cetn2
|
centrin, EF-hand protein, 2 |
| chr16_+_13818500 | 0.14 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr14_-_17622080 | 0.14 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr20_-_9436521 | 0.14 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr7_+_25059845 | 0.14 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr2_+_51183320 | 0.14 |
ENSDART00000167430
|
lrrc24
|
leucine rich repeat containing 24 |
| chr13_-_33170733 | 0.14 |
ENSDART00000057382
|
fbln5
|
fibulin 5 |
| chr23_+_6795709 | 0.14 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr10_-_33621739 | 0.13 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr11_+_10909183 | 0.13 |
ENSDART00000064860
|
rbms1a
|
RNA binding motif, single stranded interacting protein 1a |
| chr3_+_33341640 | 0.13 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr10_-_17159761 | 0.13 |
ENSDART00000080449
|
slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr11_-_6188413 | 0.13 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr19_+_42470396 | 0.13 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr1_+_8601935 | 0.13 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr7_+_17229980 | 0.13 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr2_+_20332044 | 0.13 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr7_+_25033924 | 0.13 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr10_-_8053753 | 0.13 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr17_-_37395460 | 0.13 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr9_+_33357011 | 0.13 |
ENSDART00000088569
|
nyx
|
nyctalopin |
| chr2_-_10338759 | 0.13 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr9_+_31795343 | 0.13 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr16_+_13818743 | 0.13 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr10_-_24371312 | 0.13 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr13_-_31812394 | 0.13 |
ENSDART00000124445
|
sertad4
|
SERTA domain containing 4 |
| chr19_-_31522625 | 0.12 |
ENSDART00000158438
ENSDART00000035049 |
necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr1_-_19215336 | 0.12 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr19_-_205104 | 0.12 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr6_+_39098397 | 0.12 |
ENSDART00000003716
ENSDART00000188655 |
prss60.2
|
protease, serine, 60.2 |
| chr20_-_9462433 | 0.12 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr11_-_37997419 | 0.12 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr24_+_13316737 | 0.12 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr20_-_39271844 | 0.12 |
ENSDART00000192708
|
clu
|
clusterin |
| chr24_+_2519761 | 0.12 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr1_+_12767318 | 0.12 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr3_-_34337969 | 0.12 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr6_-_41229787 | 0.12 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr23_+_20563779 | 0.12 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr6_-_38419318 | 0.12 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr13_-_32635859 | 0.12 |
ENSDART00000146249
ENSDART00000145395 ENSDART00000148040 ENSDART00000100650 |
matn3b
|
matrilin 3b |
| chr11_-_37509001 | 0.12 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr21_+_13366353 | 0.12 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr15_-_23376541 | 0.11 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr20_-_38617766 | 0.11 |
ENSDART00000050474
|
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr13_+_25449681 | 0.11 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr8_+_7144066 | 0.11 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr15_-_34213898 | 0.11 |
ENSDART00000191945
ENSDART00000186089 |
etv1
|
ets variant 1 |
| chr21_+_28958471 | 0.11 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr23_-_20051369 | 0.11 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr12_-_28881638 | 0.11 |
ENSDART00000148459
ENSDART00000039667 ENSDART00000148668 ENSDART00000136593 ENSDART00000139923 ENSDART00000148912 |
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr5_+_36611128 | 0.11 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr9_-_12424791 | 0.11 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr1_+_40023640 | 0.11 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr3_-_23406964 | 0.11 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr20_-_14925281 | 0.11 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr3_-_39695856 | 0.11 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr10_-_8053385 | 0.11 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr21_+_21655755 | 0.11 |
ENSDART00000079524
|
or125-8
|
odorant receptor, family E, subfamily 125, member 8 |
| chr14_-_33821515 | 0.11 |
ENSDART00000173218
|
vimr2
|
vimentin-related 2 |
| chr7_-_69521481 | 0.10 |
ENSDART00000148465
|
slc1a1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr22_-_13851297 | 0.10 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr10_-_8046764 | 0.10 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr6_-_38816500 | 0.10 |
ENSDART00000190866
ENSDART00000104124 |
cnga3a
|
cyclic nucleotide gated channel alpha 3a |
| chr15_-_22074315 | 0.10 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr19_-_20482261 | 0.10 |
ENSDART00000056205
|
satb1a
|
SATB homeobox 1a |
| chr24_+_5208171 | 0.10 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr23_-_44233408 | 0.10 |
ENSDART00000149318
ENSDART00000085528 |
zgc:158659
|
zgc:158659 |
| chr20_-_35040041 | 0.10 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr4_+_11384891 | 0.10 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr22_+_661505 | 0.10 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr20_-_40754794 | 0.10 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr22_-_24818066 | 0.10 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr22_+_35068046 | 0.10 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr8_-_14126646 | 0.10 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr14_-_49133426 | 0.10 |
ENSDART00000042421
|
ppp2ca
|
protein phosphatase 2, catalytic subunit, alpha isozyme |
| chr17_+_43032529 | 0.10 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr1_+_50538839 | 0.10 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr24_-_26399623 | 0.10 |
ENSDART00000112317
|
zgc:194621
|
zgc:194621 |
| chr14_-_3381303 | 0.10 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr10_+_16584382 | 0.10 |
ENSDART00000112039
|
CR790388.1
|
|
| chr23_-_26522760 | 0.10 |
ENSDART00000142417
ENSDART00000135606 ENSDART00000122668 |
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr16_+_34531486 | 0.10 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr1_+_34181581 | 0.10 |
ENSDART00000146042
|
epha6
|
eph receptor A6 |
| chr4_+_31405646 | 0.10 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr16_-_13623928 | 0.10 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr19_-_31402429 | 0.10 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr23_+_5977965 | 0.10 |
ENSDART00000115403
ENSDART00000183147 |
NAV1 (1 of many)
|
neuron navigator 1 |
| chr9_+_54417141 | 0.10 |
ENSDART00000056810
|
drd1b
|
dopamine receptor D1b |
| chr20_+_40457599 | 0.10 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr1_+_52392511 | 0.10 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr25_-_20378721 | 0.10 |
ENSDART00000181707
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr15_-_2184638 | 0.10 |
ENSDART00000135460
|
shox2
|
short stature homeobox 2 |
| chr22_-_15010688 | 0.10 |
ENSDART00000139892
|
elfn2a
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx6.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046351 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.4 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.8 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.2 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.2 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.0 | 0.1 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.2 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0043090 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.0 | 0.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.0 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.2 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.4 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.1 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.0 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0001996 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0043679 | axon terminus(GO:0043679) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 0.4 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.4 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.0 | 0.1 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.3 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.1 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |