Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
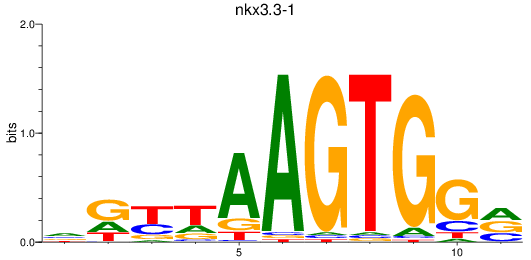
Results for nkx3.3-1
Z-value: 0.76
Transcription factors associated with nkx3.3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3.3-1
|
ENSDARG00000110589 | NK3 homeobox 3 |
Activity profile of nkx3.3-1 motif
Sorted Z-values of nkx3.3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_33308045 | 1.19 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr4_+_9467049 | 0.88 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr9_+_8380728 | 0.87 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr6_-_1820606 | 0.86 |
ENSDART00000183228
|
FO834857.1
|
|
| chr19_-_6840506 | 0.81 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr6_-_31987940 | 0.79 |
ENSDART00000132280
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr8_-_51507144 | 0.78 |
ENSDART00000024882
ENSDART00000135166 |
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr5_+_57726425 | 0.77 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr6_-_31739709 | 0.69 |
ENSDART00000087964
|
cachd1
|
cache domain containing 1 |
| chr3_-_60886984 | 0.68 |
ENSDART00000170974
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr6_-_41135215 | 0.68 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr2_-_57918314 | 0.64 |
ENSDART00000138265
|
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr13_-_50463938 | 0.63 |
ENSDART00000083857
|
ccnj
|
cyclin J |
| chr19_+_8612839 | 0.63 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr21_+_4256291 | 0.61 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 VRAC subunit Aa |
| chr23_-_29812667 | 0.61 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr13_+_33462232 | 0.60 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr2_-_57900430 | 0.60 |
ENSDART00000132245
ENSDART00000140060 |
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr4_-_5077158 | 0.60 |
ENSDART00000155915
|
ahcyl2
|
adenosylhomocysteinase-like 2 |
| chr20_+_23498255 | 0.59 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr15_+_45595385 | 0.57 |
ENSDART00000161937
ENSDART00000170214 ENSDART00000157450 |
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr25_+_28282274 | 0.57 |
ENSDART00000164502
|
aass
|
aminoadipate-semialdehyde synthase |
| chr8_+_10869183 | 0.56 |
ENSDART00000188111
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr18_-_29977431 | 0.55 |
ENSDART00000135357
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr21_-_30026359 | 0.55 |
ENSDART00000153645
|
pwwp2a
|
PWWP domain containing 2A |
| chr5_+_13870340 | 0.54 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr7_+_36467796 | 0.53 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr11_+_31680513 | 0.52 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr25_+_2361721 | 0.52 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr19_-_29302249 | 0.51 |
ENSDART00000188751
|
srfbp1
|
serum response factor binding protein 1 |
| chr5_-_57204352 | 0.51 |
ENSDART00000171252
ENSDART00000180727 |
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr6_+_58622831 | 0.50 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr5_-_54712159 | 0.50 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr7_+_20260172 | 0.50 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr10_-_7472323 | 0.48 |
ENSDART00000163702
ENSDART00000167054 ENSDART00000167706 |
nrg1
|
neuregulin 1 |
| chr12_+_29236274 | 0.48 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr16_+_3982590 | 0.47 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr21_+_15435946 | 0.47 |
ENSDART00000137491
|
si:dkey-11o15.10
|
si:dkey-11o15.10 |
| chr23_-_24542952 | 0.47 |
ENSDART00000088777
|
atp13a2
|
ATPase 13A2 |
| chr17_-_25831569 | 0.46 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr22_-_16755885 | 0.46 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr19_+_32321797 | 0.46 |
ENSDART00000167664
|
atxn1a
|
ataxin 1a |
| chr5_-_38248347 | 0.46 |
ENSDART00000084917
ENSDART00000139479 |
slc12a9
|
solute carrier family 12, member 9 |
| chr8_-_51753604 | 0.45 |
ENSDART00000007090
|
tbx16
|
T-box 16 |
| chr4_-_14642379 | 0.44 |
ENSDART00000114977
|
si:ch211-127b11.1
|
si:ch211-127b11.1 |
| chr1_-_58887610 | 0.44 |
ENSDART00000180647
|
CABZ01084501.1
|
microfibril-associated glycoprotein 4-like precursor |
| chr16_-_22585289 | 0.44 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr7_-_23777445 | 0.44 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr6_-_12275836 | 0.43 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr20_+_46741074 | 0.43 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr16_-_13612650 | 0.42 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr23_+_25172976 | 0.41 |
ENSDART00000140789
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr7_-_70283372 | 0.41 |
ENSDART00000112969
|
adgra3
|
adhesion G protein-coupled receptor A3 |
| chr11_+_27134116 | 0.41 |
ENSDART00000129736
|
hdac11
|
histone deacetylase 11 |
| chr10_-_33343244 | 0.40 |
ENSDART00000164191
|
c2cd2
|
C2 calcium-dependent domain containing 2 |
| chr23_-_26227805 | 0.40 |
ENSDART00000158082
|
BX927204.1
|
|
| chr5_-_55848511 | 0.40 |
ENSDART00000183503
|
camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr5_-_5243079 | 0.39 |
ENSDART00000130576
ENSDART00000164377 |
mvb12ba
|
multivesicular body subunit 12Ba |
| chr6_+_515181 | 0.38 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr16_+_27444098 | 0.37 |
ENSDART00000157690
|
invs
|
inversin |
| chr4_-_14649158 | 0.37 |
ENSDART00000145737
|
si:dkey-183c2.4
|
si:dkey-183c2.4 |
| chr21_+_18925318 | 0.37 |
ENSDART00000136182
|
si:ch211-222n4.2
|
si:ch211-222n4.2 |
| chr7_-_5487593 | 0.37 |
ENSDART00000136594
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr3_-_31069776 | 0.37 |
ENSDART00000167462
|
elob
|
elongin B |
| chr5_+_44846280 | 0.36 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr8_-_43689324 | 0.36 |
ENSDART00000159904
|
ep400
|
E1A binding protein p400 |
| chr23_-_17003533 | 0.35 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr16_+_38240027 | 0.35 |
ENSDART00000111081
|
prune
|
prune exopolyphosphatase |
| chr12_+_16087077 | 0.35 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr20_-_33675676 | 0.35 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr8_-_14554785 | 0.35 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr10_+_37268854 | 0.34 |
ENSDART00000131897
|
nf1b
|
neurofibromin 1b |
| chr3_+_14581643 | 0.34 |
ENSDART00000182840
|
znf653
|
zinc finger protein 653 |
| chr14_-_51014292 | 0.34 |
ENSDART00000029797
|
faf2
|
Fas associated factor family member 2 |
| chr3_+_11926030 | 0.33 |
ENSDART00000081367
|
dnaja3a
|
DnaJ (Hsp40) homolog, subfamily A, member 3A |
| chr12_-_45304971 | 0.33 |
ENSDART00000186537
ENSDART00000126405 |
fdxr
|
ferredoxin reductase |
| chr4_-_8030583 | 0.32 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr8_-_4031121 | 0.32 |
ENSDART00000169474
ENSDART00000163754 |
mtmr3
|
myotubularin related protein 3 |
| chr8_-_8446430 | 0.32 |
ENSDART00000137382
|
cdk16
|
cyclin-dependent kinase 16 |
| chr16_+_6750289 | 0.31 |
ENSDART00000167736
|
znf236
|
zinc finger protein 236 |
| chr13_+_23093743 | 0.31 |
ENSDART00000148034
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr18_+_22174630 | 0.31 |
ENSDART00000089549
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr16_-_12097558 | 0.31 |
ENSDART00000123142
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr22_-_27113332 | 0.31 |
ENSDART00000146951
ENSDART00000178855 |
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr25_+_33033633 | 0.31 |
ENSDART00000192336
|
tln2b
|
talin 2b |
| chr11_-_5953636 | 0.30 |
ENSDART00000140960
ENSDART00000123601 |
dda1
|
DET1 and DDB1 associated 1 |
| chr8_+_8712446 | 0.30 |
ENSDART00000158674
|
elk1
|
ELK1, member of ETS oncogene family |
| chr5_-_57820873 | 0.30 |
ENSDART00000089961
|
sik2a
|
salt-inducible kinase 2a |
| chr17_-_16069905 | 0.29 |
ENSDART00000110383
|
map7a
|
microtubule-associated protein 7a |
| chr17_+_21817859 | 0.29 |
ENSDART00000143832
ENSDART00000141462 |
ikzf5
|
IKAROS family zinc finger 5 |
| chr24_-_41195068 | 0.29 |
ENSDART00000121592
|
acvr2ba
|
activin A receptor type 2Ba |
| chr5_+_29652198 | 0.29 |
ENSDART00000184083
|
tsc1a
|
TSC complex subunit 1a |
| chr5_-_8907819 | 0.29 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr16_-_26132122 | 0.29 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr5_-_29122834 | 0.29 |
ENSDART00000087197
|
whrnb
|
whirlin b |
| chr22_+_7480465 | 0.29 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr3_-_52899394 | 0.29 |
ENSDART00000128223
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr19_-_43757568 | 0.29 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr22_+_9922301 | 0.28 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr16_+_46725087 | 0.28 |
ENSDART00000008920
|
rab11al
|
RAB11a, member RAS oncogene family, like |
| chr14_-_36763302 | 0.28 |
ENSDART00000074786
|
ctso
|
cathepsin O |
| chr11_+_19370717 | 0.28 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr17_+_23554932 | 0.28 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr20_-_53078607 | 0.28 |
ENSDART00000163494
ENSDART00000191730 |
CABZ01066813.1
|
|
| chr5_-_23783739 | 0.28 |
ENSDART00000139502
|
GBGT1 (1 of many)
|
si:ch211-287c22.1 |
| chr19_+_791538 | 0.28 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr18_+_44566863 | 0.28 |
ENSDART00000176084
|
dhx34
|
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
| chr20_-_28698172 | 0.28 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr22_+_32228882 | 0.27 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr24_-_25166720 | 0.27 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr11_+_19370447 | 0.26 |
ENSDART00000186154
|
prickle2b
|
prickle homolog 2b |
| chr1_-_26045560 | 0.26 |
ENSDART00000172737
ENSDART00000076120 ENSDART00000193593 |
gpank1
|
G patch domain and ankyrin repeats 1 |
| chr11_-_2221159 | 0.26 |
ENSDART00000189292
|
smug1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr7_-_30272871 | 0.25 |
ENSDART00000099586
|
zgc:162945
|
zgc:162945 |
| chr25_+_7299488 | 0.25 |
ENSDART00000184836
|
hmg20a
|
high mobility group 20A |
| chr22_-_31020690 | 0.25 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr1_+_55095314 | 0.24 |
ENSDART00000132727
|
aftpha
|
aftiphilin a |
| chr1_+_18863060 | 0.24 |
ENSDART00000139241
|
rnf38
|
ring finger protein 38 |
| chr20_+_27298783 | 0.24 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr11_+_19271557 | 0.23 |
ENSDART00000190559
|
prickle2b
|
prickle homolog 2b |
| chr19_+_40248697 | 0.23 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr17_+_21818093 | 0.23 |
ENSDART00000125335
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr3_-_34753605 | 0.23 |
ENSDART00000000160
|
thraa
|
thyroid hormone receptor alpha a |
| chr5_-_29122615 | 0.23 |
ENSDART00000144802
|
whrnb
|
whirlin b |
| chr14_+_49152341 | 0.22 |
ENSDART00000084114
|
nsd1a
|
nuclear receptor binding SET domain protein 1a |
| chr10_-_38468847 | 0.22 |
ENSDART00000133914
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr9_+_51655636 | 0.22 |
ENSDART00000169908
|
RBMS1 (1 of many)
|
RNA binding motif single stranded interacting protein 1 |
| chr19_-_35155722 | 0.22 |
ENSDART00000151924
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr5_-_26765188 | 0.22 |
ENSDART00000029450
|
rnf181
|
ring finger protein 181 |
| chr6_+_41808673 | 0.22 |
ENSDART00000038163
|
rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr9_-_1604601 | 0.22 |
ENSDART00000143130
|
agps
|
alkylglycerone phosphate synthase |
| chr10_+_44641599 | 0.22 |
ENSDART00000172128
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr18_+_26829362 | 0.22 |
ENSDART00000132728
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr8_+_53051701 | 0.21 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr15_-_20709289 | 0.21 |
ENSDART00000136767
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr23_-_18568522 | 0.21 |
ENSDART00000004655
|
sephs2
|
selenophosphate synthetase 2 |
| chr22_-_34551568 | 0.21 |
ENSDART00000148147
|
rnf123
|
ring finger protein 123 |
| chr1_-_524433 | 0.21 |
ENSDART00000147610
|
si:ch73-41e3.7
|
si:ch73-41e3.7 |
| chr13_+_10023256 | 0.21 |
ENSDART00000110035
|
srbd1
|
S1 RNA binding domain 1 |
| chr18_-_22094102 | 0.21 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr1_+_55643198 | 0.20 |
ENSDART00000060693
|
adgre7
|
adhesion G protein-coupled receptor E7 |
| chr23_+_12840080 | 0.20 |
ENSDART00000081016
ENSDART00000121697 |
smc1al
|
structural maintenance of chromosomes 1A, like |
| chr5_-_12063381 | 0.20 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr23_-_21535040 | 0.20 |
ENSDART00000010647
|
rcc2
|
regulator of chromosome condensation 2 |
| chr10_-_38456382 | 0.20 |
ENSDART00000182129
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr15_-_24676211 | 0.20 |
ENSDART00000153502
ENSDART00000100756 |
tmem199
|
transmembrane protein 199 |
| chr14_-_31814149 | 0.19 |
ENSDART00000173393
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr2_+_55916911 | 0.19 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr9_-_34882516 | 0.19 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr23_+_36601984 | 0.18 |
ENSDART00000128598
|
igfbp6b
|
insulin-like growth factor binding protein 6b |
| chr24_-_11057305 | 0.18 |
ENSDART00000186494
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr7_-_40578733 | 0.18 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr5_-_26764880 | 0.18 |
ENSDART00000140392
ENSDART00000134728 |
rnf181
|
ring finger protein 181 |
| chr23_-_36418708 | 0.18 |
ENSDART00000132273
|
znf740b
|
zinc finger protein 740b |
| chr14_+_21722235 | 0.18 |
ENSDART00000183667
|
stx3a
|
syntaxin 3A |
| chr6_+_3280939 | 0.17 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr21_+_34814444 | 0.17 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr25_+_11456696 | 0.17 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr25_-_37284370 | 0.17 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr16_+_20934353 | 0.17 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr4_-_2059233 | 0.17 |
ENSDART00000188177
ENSDART00000129521 ENSDART00000082289 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr21_+_10021823 | 0.17 |
ENSDART00000163995
|
herc7
|
hect domain and RLD 7 |
| chr5_+_26765275 | 0.17 |
ENSDART00000144169
|
si:ch211-102c2.8
|
si:ch211-102c2.8 |
| chr15_+_20281305 | 0.16 |
ENSDART00000155065
|
plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr25_-_11016675 | 0.16 |
ENSDART00000099572
|
mespab
|
mesoderm posterior ab |
| chr1_-_30568225 | 0.16 |
ENSDART00000144297
ENSDART00000164204 |
ubac2
|
UBA domain containing 2 |
| chr6_+_46431848 | 0.16 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr1_-_49950643 | 0.15 |
ENSDART00000138301
|
sgms2
|
sphingomyelin synthase 2 |
| chr21_-_38031038 | 0.15 |
ENSDART00000179483
ENSDART00000076238 |
rbm41
|
RNA binding motif protein 41 |
| chr1_-_54706039 | 0.15 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr15_+_42285643 | 0.15 |
ENSDART00000152731
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr15_+_15771418 | 0.15 |
ENSDART00000153831
|
si:ch211-33e4.3
|
si:ch211-33e4.3 |
| chr4_-_10835620 | 0.14 |
ENSDART00000150739
|
ppfibp1a
|
PTPRF interacting protein, binding protein 1a (liprin beta 1) |
| chr3_-_18384501 | 0.14 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr13_+_18371208 | 0.14 |
ENSDART00000138172
|
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr21_+_26612777 | 0.14 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr9_+_15893093 | 0.13 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr19_+_30990815 | 0.13 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr12_+_17754859 | 0.13 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr8_-_25814263 | 0.13 |
ENSDART00000143397
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr13_-_35892051 | 0.12 |
ENSDART00000145884
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr6_-_59381391 | 0.12 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr16_+_14710436 | 0.12 |
ENSDART00000027982
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr17_-_51262430 | 0.11 |
ENSDART00000163222
|
trappc12
|
trafficking protein particle complex 12 |
| chr8_-_18229169 | 0.11 |
ENSDART00000131764
ENSDART00000143036 ENSDART00000145986 |
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr15_-_43873005 | 0.11 |
ENSDART00000190326
|
NOX4
|
NADPH oxidase 4 |
| chr7_-_59311165 | 0.11 |
ENSDART00000171105
|
m1ap
|
meiosis 1 associated protein |
| chr6_-_52235118 | 0.11 |
ENSDART00000191243
|
tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr22_+_724639 | 0.11 |
ENSDART00000105323
|
zgc:162255
|
zgc:162255 |
| chr3_-_21010979 | 0.11 |
ENSDART00000109578
|
fam117aa
|
family with sequence similarity 117, member Aa |
| chr9_-_43073960 | 0.10 |
ENSDART00000059460
|
ttn.2
|
titin, tandem duplicate 2 |
| chr10_+_40660772 | 0.10 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr21_-_2158298 | 0.10 |
ENSDART00000182199
|
gb:ai877918
|
expressed sequence AI877918 |
| chr17_+_38447473 | 0.10 |
ENSDART00000149007
|
cdan1
|
codanin 1 |
| chr21_-_21527983 | 0.10 |
ENSDART00000014254
|
diablob
|
diablo, IAP-binding mitochondrial protein b |
| chr22_+_1330477 | 0.10 |
ENSDART00000157567
|
CU207221.2
|
|
| chr12_+_8074343 | 0.10 |
ENSDART00000124084
|
cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr8_+_25254435 | 0.10 |
ENSDART00000143554
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr2_+_30463825 | 0.10 |
ENSDART00000092356
|
neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr22_-_36750589 | 0.10 |
ENSDART00000010824
|
acy1
|
aminoacylase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3.3-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.2 | 0.6 | GO:0033512 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.2 | 0.5 | GO:0060959 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.2 | 0.5 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) |
| 0.1 | 0.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.6 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.5 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.5 | GO:0097094 | regulation of osteoblast proliferation(GO:0033688) craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.4 | GO:0019075 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.1 | 0.4 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.8 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.2 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.5 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.3 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.5 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.3 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.1 | 0.2 | GO:0046833 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.4 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.5 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.4 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.6 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.2 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.0 | 0.5 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.5 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.2 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.8 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.4 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.8 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.3 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.3 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.4 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.0 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.5 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.4 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.0 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 1.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.5 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.6 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.5 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.6 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.6 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | Genes involved in Activation of NMDA receptor upon glutamate binding and postsynaptic events |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |