Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
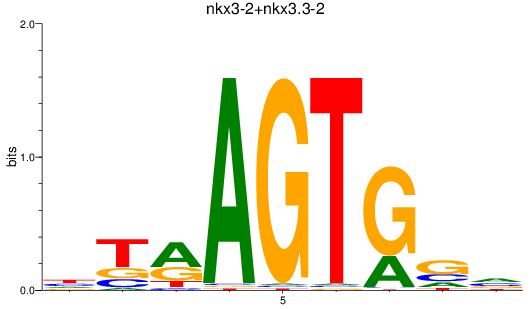
Results for nkx3-2+nkx3.3-2
Z-value: 0.40
Transcription factors associated with nkx3-2+nkx3.3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3-2
|
ENSDARG00000037639 | NK3 homeobox 2 |
|
nkx3.3-2
|
ENSDARG00000069327 | NK3 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx3.2 | dr11_v1_chr14_-_215051_215051 | -0.60 | 8.9e-02 | Click! |
| nkx3.3 | dr11_v1_chr13_-_40401870_40401870 | -0.44 | 2.3e-01 | Click! |
Activity profile of nkx3-2+nkx3.3-2 motif
Sorted Z-values of nkx3-2+nkx3.3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_37929135 | 0.67 |
ENSDART00000157550
|
si:dkey-238d18.15
|
si:dkey-238d18.15 |
| chr11_+_14333441 | 0.32 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr12_+_17754859 | 0.30 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr22_+_9922301 | 0.29 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr12_-_13729263 | 0.28 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr17_+_51746830 | 0.25 |
ENSDART00000184230
|
odc1
|
ornithine decarboxylase 1 |
| chr20_+_25568694 | 0.23 |
ENSDART00000063107
ENSDART00000063128 |
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr9_-_9225980 | 0.23 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr15_+_888704 | 0.22 |
ENSDART00000182796
|
si:dkey-7i4.9
|
si:dkey-7i4.9 |
| chr8_+_25254435 | 0.21 |
ENSDART00000143554
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr21_+_18353703 | 0.19 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr7_+_44715224 | 0.19 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr4_+_45357558 | 0.19 |
ENSDART00000150769
|
si:ch211-162i8.5
|
si:ch211-162i8.5 |
| chr1_-_51710225 | 0.19 |
ENSDART00000057601
ENSDART00000152745 |
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr24_+_17005647 | 0.18 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr1_+_7988052 | 0.18 |
ENSDART00000167552
|
CR855320.2
|
|
| chr23_-_33558161 | 0.17 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr7_-_6441865 | 0.17 |
ENSDART00000172831
|
hist1h2a10
|
histone cluster 1 H2A family member 10 |
| chr18_-_22094102 | 0.17 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr25_+_16356083 | 0.16 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr9_-_49829685 | 0.16 |
ENSDART00000162271
|
ttc21b
|
tetratricopeptide repeat domain 21B |
| chr21_+_34814444 | 0.16 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr20_-_25643667 | 0.16 |
ENSDART00000137457
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr22_+_2830703 | 0.16 |
ENSDART00000145463
ENSDART00000144785 |
si:dkey-20i20.8
|
si:dkey-20i20.8 |
| chr12_-_16636627 | 0.15 |
ENSDART00000128811
|
si:dkey-239j18.3
|
si:dkey-239j18.3 |
| chr7_+_20917966 | 0.14 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr18_+_16943911 | 0.14 |
ENSDART00000157609
|
si:dkey-8l13.5
|
si:dkey-8l13.5 |
| chr8_+_20455134 | 0.13 |
ENSDART00000079618
|
rexo1
|
REX1, RNA exonuclease 1 homolog |
| chr15_-_35246742 | 0.12 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr13_+_12606821 | 0.12 |
ENSDART00000140096
ENSDART00000145136 |
metap1
|
methionyl aminopeptidase 1 |
| chr18_-_26781616 | 0.11 |
ENSDART00000136776
ENSDART00000076484 |
kti12
|
KTI12 chromatin associated homolog |
| chr6_-_39198912 | 0.10 |
ENSDART00000077938
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr22_-_27113332 | 0.10 |
ENSDART00000146951
ENSDART00000178855 |
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr17_-_21278582 | 0.10 |
ENSDART00000157518
|
hspa12a
|
heat shock protein 12A |
| chr17_+_31742923 | 0.10 |
ENSDART00000086696
|
arhgap5
|
Rho GTPase activating protein 5 |
| chr19_+_43604643 | 0.10 |
ENSDART00000151168
|
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr10_-_29831944 | 0.09 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr18_-_20458412 | 0.09 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr4_+_17327704 | 0.09 |
ENSDART00000016075
ENSDART00000133160 |
nup37
|
nucleoporin 37 |
| chr16_+_26612401 | 0.09 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr15_+_5360407 | 0.09 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr23_+_12840080 | 0.09 |
ENSDART00000081016
ENSDART00000121697 |
smc1al
|
structural maintenance of chromosomes 1A, like |
| chr4_+_13931578 | 0.09 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr16_+_28994709 | 0.09 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr17_-_21278846 | 0.09 |
ENSDART00000181356
|
hspa12a
|
heat shock protein 12A |
| chr21_-_38031038 | 0.09 |
ENSDART00000179483
ENSDART00000076238 |
rbm41
|
RNA binding motif protein 41 |
| chr15_+_24676905 | 0.08 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr3_-_53559408 | 0.08 |
ENSDART00000073930
|
notch3
|
notch 3 |
| chr1_-_48933 | 0.08 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr21_-_2348838 | 0.08 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr4_-_13931508 | 0.08 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr24_+_21676921 | 0.08 |
ENSDART00000066677
|
si:ch211-140b10.6
|
si:ch211-140b10.6 |
| chr2_+_42135719 | 0.07 |
ENSDART00000008268
|
cyp7b1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr4_+_72668095 | 0.07 |
ENSDART00000182282
ENSDART00000162637 |
CABZ01054391.1
|
|
| chr3_-_18384501 | 0.07 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr23_+_25292147 | 0.07 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr18_+_22220656 | 0.07 |
ENSDART00000191862
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr2_+_55916911 | 0.07 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr4_+_71018579 | 0.07 |
ENSDART00000186727
|
si:dkeyp-80d11.10
|
si:dkeyp-80d11.10 |
| chr3_-_53559581 | 0.07 |
ENSDART00000183499
|
notch3
|
notch 3 |
| chr22_-_20386982 | 0.06 |
ENSDART00000089015
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr16_+_27444098 | 0.06 |
ENSDART00000157690
|
invs
|
inversin |
| chr11_-_1948784 | 0.06 |
ENSDART00000082475
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr12_+_26706745 | 0.06 |
ENSDART00000141401
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr11_-_12634017 | 0.06 |
ENSDART00000158286
ENSDART00000193090 |
CR450764.1
|
|
| chr4_-_5077158 | 0.06 |
ENSDART00000155915
|
ahcyl2
|
adenosylhomocysteinase-like 2 |
| chrM_+_9735 | 0.06 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr8_+_24281512 | 0.06 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr24_-_31223232 | 0.06 |
ENSDART00000164155
|
alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr5_-_68779747 | 0.06 |
ENSDART00000192636
ENSDART00000188039 |
mepce
|
methylphosphate capping enzyme |
| chr3_-_16413606 | 0.05 |
ENSDART00000127309
ENSDART00000017172 ENSDART00000136465 |
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr6_+_13201358 | 0.05 |
ENSDART00000190290
|
CT009620.1
|
|
| chr16_-_16522013 | 0.05 |
ENSDART00000160602
|
nbeal2
|
neurobeachin-like 2 |
| chr14_-_1200854 | 0.05 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr20_-_30377221 | 0.05 |
ENSDART00000126229
|
rps7
|
ribosomal protein S7 |
| chr11_-_5953636 | 0.05 |
ENSDART00000140960
ENSDART00000123601 |
dda1
|
DET1 and DDB1 associated 1 |
| chr20_-_438646 | 0.04 |
ENSDART00000009196
|
ufl1
|
UFM1-specific ligase 1 |
| chr19_+_19989380 | 0.04 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr19_-_3777217 | 0.04 |
ENSDART00000160510
|
si:dkey-206d17.15
|
si:dkey-206d17.15 |
| chr4_-_75812937 | 0.04 |
ENSDART00000125096
|
si:ch211-203c5.3
|
si:ch211-203c5.3 |
| chr14_-_26392475 | 0.04 |
ENSDART00000170614
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr22_+_19188809 | 0.04 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr3_+_19446997 | 0.04 |
ENSDART00000079352
|
zgc:123297
|
zgc:123297 |
| chr21_+_4388489 | 0.04 |
ENSDART00000144555
|
HTRA2 (1 of many)
|
si:dkey-84o3.3 |
| chr18_-_29977431 | 0.03 |
ENSDART00000135357
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr22_-_23545307 | 0.03 |
ENSDART00000166915
|
crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr7_-_4843401 | 0.03 |
ENSDART00000137489
|
si:dkey-83f18.8
|
si:dkey-83f18.8 |
| chr17_+_6563307 | 0.03 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr15_-_36347858 | 0.03 |
ENSDART00000155274
ENSDART00000157936 |
si:dkey-23k10.2
|
si:dkey-23k10.2 |
| chr13_+_15581270 | 0.03 |
ENSDART00000189880
ENSDART00000190067 ENSDART00000041293 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr8_-_4971205 | 0.03 |
ENSDART00000184250
|
tmem230b
|
transmembrane protein 230b |
| chr2_-_59157790 | 0.03 |
ENSDART00000192303
ENSDART00000159362 |
ftr32
|
finTRIM family, member 32 |
| chr19_+_32855139 | 0.03 |
ENSDART00000052082
|
rpl30
|
ribosomal protein L30 |
| chr15_+_15173611 | 0.03 |
ENSDART00000155267
|
si:ch211-149e23.4
|
si:ch211-149e23.4 |
| chr21_+_4403155 | 0.03 |
ENSDART00000161623
|
HTRA2 (1 of many)
|
si:dkey-84o3.3 |
| chr10_+_24048883 | 0.02 |
ENSDART00000149265
|
gbe1a
|
glucan (1,4-alpha-), branching enzyme 1a |
| chr10_+_24048663 | 0.02 |
ENSDART00000184504
ENSDART00000186955 |
gbe1a
|
glucan (1,4-alpha-), branching enzyme 1a |
| chr19_+_30990129 | 0.02 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr4_-_71436560 | 0.02 |
ENSDART00000166884
|
si:dkey-82i20.1
|
si:dkey-82i20.1 |
| chr8_+_28547687 | 0.02 |
ENSDART00000110291
|
srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr5_-_8907819 | 0.02 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr1_-_30039331 | 0.01 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr22_-_26514386 | 0.01 |
ENSDART00000125628
|
CR759791.1
|
|
| chr18_+_22174630 | 0.01 |
ENSDART00000089549
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr4_+_71382288 | 0.01 |
ENSDART00000181926
|
si:ch211-76m11.8
|
si:ch211-76m11.8 |
| chr16_-_31933740 | 0.00 |
ENSDART00000125411
|
si:ch1073-90m23.1
|
si:ch1073-90m23.1 |
| chr12_+_13404784 | 0.00 |
ENSDART00000167977
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr18_-_27858123 | 0.00 |
ENSDART00000142068
|
iqcg
|
IQ motif containing G |
| chr5_+_61843752 | 0.00 |
ENSDART00000130940
|
CR759879.1
|
Danio rerio interferon-induced protein with tetratricopeptide repeats 5 (LOC572297), mRNA. |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3-2+nkx3.3-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.1 | 0.2 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 0.2 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.1 | GO:1990173 | positive regulation of protein localization to nucleus(GO:1900182) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.2 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.1 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.0 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |