Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
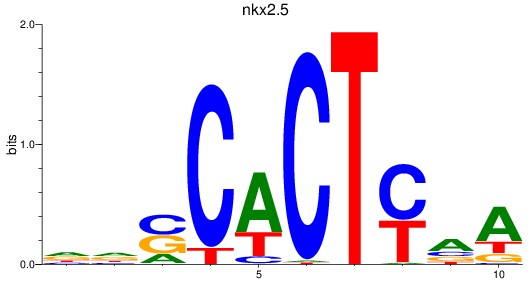
Results for nkx2.5
Z-value: 1.02
Transcription factors associated with nkx2.5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.5
|
ENSDARG00000018004 | NK2 homeobox 5 |
|
nkx2.5
|
ENSDARG00000116714 | NK2 homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.5 | dr11_v1_chr14_+_24241241_24241241 | -0.77 | 1.5e-02 | Click! |
Activity profile of nkx2.5 motif
Sorted Z-values of nkx2.5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_25236888 | 2.41 |
ENSDART00000037286
|
zgc:172218
|
zgc:172218 |
| chr12_-_13729263 | 1.70 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr4_-_77130289 | 1.68 |
ENSDART00000174380
|
CU467646.7
|
|
| chr4_-_77125693 | 1.66 |
ENSDART00000174256
|
CU467646.3
|
|
| chr4_-_77135076 | 1.56 |
ENSDART00000174184
|
zgc:173770
|
zgc:173770 |
| chr22_+_25242322 | 1.51 |
ENSDART00000134628
|
si:ch211-226h8.8
|
si:ch211-226h8.8 |
| chr9_+_8396755 | 1.41 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr1_-_6028876 | 1.23 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr6_+_515181 | 1.20 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr22_+_25236657 | 1.17 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr6_-_1820606 | 1.17 |
ENSDART00000183228
|
FO834857.1
|
|
| chr19_-_2115040 | 1.13 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr3_+_32571929 | 1.08 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr16_+_26612401 | 1.01 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr13_+_23093743 | 0.98 |
ENSDART00000148034
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr6_-_24384654 | 0.96 |
ENSDART00000164723
|
brdt
|
bromodomain, testis-specific |
| chr8_+_3431671 | 0.96 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr24_+_15655233 | 0.95 |
ENSDART00000143160
|
fbxo15
|
F-box protein 15 |
| chr6_+_58622831 | 0.94 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr16_-_22585289 | 0.89 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr24_+_10039165 | 0.89 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr20_+_42918755 | 0.88 |
ENSDART00000134855
|
efr3bb
|
EFR3 homolog Bb (S. cerevisiae) |
| chr8_+_25893071 | 0.88 |
ENSDART00000078161
|
tmem115
|
transmembrane protein 115 |
| chr10_-_38498072 | 0.84 |
ENSDART00000111234
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr12_+_33320884 | 0.83 |
ENSDART00000188988
|
csnk1db
|
casein kinase 1, delta b |
| chr5_+_24543862 | 0.81 |
ENSDART00000029699
|
atp6v0a2b
|
ATPase H+ transporting V0 subunit a2b |
| chr23_+_30736895 | 0.81 |
ENSDART00000042944
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr6_-_12900154 | 0.79 |
ENSDART00000080408
ENSDART00000150887 |
ical1
|
islet cell autoantigen 1-like |
| chr15_-_26568278 | 0.76 |
ENSDART00000182609
|
wdr81
|
WD repeat domain 81 |
| chr6_-_7726849 | 0.75 |
ENSDART00000151511
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr3_-_10582384 | 0.75 |
ENSDART00000048095
ENSDART00000155152 |
elac2
|
elaC ribonuclease Z 2 |
| chr14_+_31493306 | 0.71 |
ENSDART00000138341
|
phf6
|
PHD finger protein 6 |
| chr18_+_17534627 | 0.71 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr8_-_4100365 | 0.70 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr14_+_31493119 | 0.69 |
ENSDART00000006463
|
phf6
|
PHD finger protein 6 |
| chr18_-_399554 | 0.69 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr3_+_14463941 | 0.66 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr18_-_29977431 | 0.66 |
ENSDART00000135357
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr3_+_22905341 | 0.66 |
ENSDART00000111435
|
hdac5
|
histone deacetylase 5 |
| chr2_+_30513887 | 0.65 |
ENSDART00000137048
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr17_+_6538733 | 0.64 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr8_+_9715010 | 0.64 |
ENSDART00000139414
|
gripap1
|
GRIP1 associated protein 1 |
| chr6_-_6673813 | 0.64 |
ENSDART00000150995
|
si:dkey-261j15.2
|
si:dkey-261j15.2 |
| chr3_+_35812040 | 0.63 |
ENSDART00000075903
ENSDART00000147712 |
crlf3
|
cytokine receptor-like factor 3 |
| chr22_+_15973122 | 0.62 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr1_+_44053641 | 0.60 |
ENSDART00000124873
|
si:ch73-109d9.4
|
si:ch73-109d9.4 |
| chr8_-_22558773 | 0.60 |
ENSDART00000074309
|
porcnl
|
porcupine O-acyltransferase like |
| chr3_+_14581643 | 0.60 |
ENSDART00000182840
|
znf653
|
zinc finger protein 653 |
| chr16_+_38240027 | 0.59 |
ENSDART00000111081
|
prune
|
prune exopolyphosphatase |
| chr12_+_32292564 | 0.58 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr7_-_48733662 | 0.58 |
ENSDART00000191675
|
traf6
|
TNF receptor-associated factor 6 |
| chr9_+_38737924 | 0.57 |
ENSDART00000147652
|
kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr23_-_27479558 | 0.57 |
ENSDART00000013563
|
atf7a
|
activating transcription factor 7a |
| chr8_+_11471350 | 0.56 |
ENSDART00000092355
ENSDART00000136184 |
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr7_-_23777445 | 0.56 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr13_-_28610965 | 0.55 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr23_+_7710721 | 0.55 |
ENSDART00000186852
ENSDART00000161193 |
kif3b
|
kinesin family member 3B |
| chr13_-_50614639 | 0.54 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr12_-_45304971 | 0.54 |
ENSDART00000186537
ENSDART00000126405 |
fdxr
|
ferredoxin reductase |
| chr18_+_3064237 | 0.54 |
ENSDART00000161848
|
ints4
|
integrator complex subunit 4 |
| chr8_+_25962833 | 0.54 |
ENSDART00000086583
|
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr16_-_12097558 | 0.54 |
ENSDART00000123142
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr20_+_14789305 | 0.54 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr24_-_23839647 | 0.53 |
ENSDART00000125190
|
rrs1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr5_+_47975758 | 0.52 |
ENSDART00000097429
|
BX470189.1
|
|
| chr12_+_5102670 | 0.52 |
ENSDART00000166600
|
cep55l
|
centrosomal protein 55 like |
| chr20_+_23498255 | 0.52 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr20_-_25643667 | 0.52 |
ENSDART00000137457
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr23_+_7710447 | 0.51 |
ENSDART00000168199
|
kif3b
|
kinesin family member 3B |
| chr20_-_28698172 | 0.51 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr18_-_46183462 | 0.49 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr22_+_2229964 | 0.49 |
ENSDART00000112582
|
znf1161
|
zinc finger protein 1161 |
| chr11_-_18017287 | 0.49 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr4_-_39111612 | 0.48 |
ENSDART00000150394
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr3_-_26805455 | 0.48 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr8_+_21114338 | 0.48 |
ENSDART00000002186
|
uck2a
|
uridine-cytidine kinase 2a |
| chr23_-_14769523 | 0.48 |
ENSDART00000054909
|
gss
|
glutathione synthetase |
| chr5_-_29122834 | 0.47 |
ENSDART00000087197
|
whrnb
|
whirlin b |
| chr9_-_13871935 | 0.47 |
ENSDART00000146597
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr16_-_8132742 | 0.47 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr1_+_16621345 | 0.46 |
ENSDART00000149026
|
pcm1
|
pericentriolar material 1 |
| chr13_-_32899322 | 0.46 |
ENSDART00000133882
|
rock2a
|
rho-associated, coiled-coil containing protein kinase 2a |
| chr21_+_34814444 | 0.46 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr8_-_20243389 | 0.45 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr17_+_13088594 | 0.45 |
ENSDART00000193207
|
gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr15_-_36727462 | 0.45 |
ENSDART00000085971
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr25_+_16194979 | 0.45 |
ENSDART00000185592
ENSDART00000158582 ENSDART00000161109 ENSDART00000139013 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr15_-_17138640 | 0.44 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr15_+_24676905 | 0.44 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr5_+_18047111 | 0.44 |
ENSDART00000132164
|
hira
|
histone cell cycle regulator a |
| chr11_-_27537593 | 0.43 |
ENSDART00000173444
ENSDART00000172895 ENSDART00000088177 |
ptpdc1a
|
protein tyrosine phosphatase domain containing 1a |
| chr24_-_26945390 | 0.43 |
ENSDART00000123354
|
msl2b
|
male-specific lethal 2 homolog b (Drosophila) |
| chr17_-_10043273 | 0.43 |
ENSDART00000156078
|
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr5_-_8907819 | 0.42 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr16_-_41787421 | 0.42 |
ENSDART00000147210
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr1_+_46598502 | 0.42 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr2_-_40890004 | 0.42 |
ENSDART00000191746
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr12_+_5251647 | 0.42 |
ENSDART00000124097
|
plce1
|
phospholipase C, epsilon 1 |
| chr20_+_34390196 | 0.42 |
ENSDART00000183596
|
trmt1l
|
tRNA methyltransferase 1-like |
| chr24_+_35183595 | 0.42 |
ENSDART00000075142
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr5_-_29122615 | 0.42 |
ENSDART00000144802
|
whrnb
|
whirlin b |
| chr4_-_4795205 | 0.42 |
ENSDART00000039313
|
zgc:162331
|
zgc:162331 |
| chr3_+_39600562 | 0.41 |
ENSDART00000134309
ENSDART00000007170 |
prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr22_+_1480988 | 0.41 |
ENSDART00000171122
|
si:ch211-255f4.2
|
si:ch211-255f4.2 |
| chr5_+_2002804 | 0.41 |
ENSDART00000064088
|
vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr4_-_13255700 | 0.41 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr22_-_27113332 | 0.40 |
ENSDART00000146951
ENSDART00000178855 |
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr9_+_17862858 | 0.40 |
ENSDART00000166566
|
dgkh
|
diacylglycerol kinase, eta |
| chr25_-_34723937 | 0.39 |
ENSDART00000187286
ENSDART00000157021 |
nup160
|
nucleoporin 160 |
| chr13_-_31346392 | 0.38 |
ENSDART00000134343
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr22_+_1421212 | 0.38 |
ENSDART00000161813
|
zgc:101130
|
zgc:101130 |
| chr7_-_67248829 | 0.38 |
ENSDART00000192442
|
znf143a
|
zinc finger protein 143a |
| chr14_+_21828993 | 0.38 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr10_+_1052591 | 0.37 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr9_+_2452672 | 0.37 |
ENSDART00000193993
|
chn1
|
chimerin 1 |
| chr11_-_29082175 | 0.37 |
ENSDART00000123245
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr11_-_34232906 | 0.36 |
ENSDART00000162150
|
lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr20_-_26588736 | 0.36 |
ENSDART00000134337
|
exoc2
|
exocyst complex component 2 |
| chr2_-_37280617 | 0.36 |
ENSDART00000190458
|
nadkb
|
NAD kinase b |
| chr5_-_24543526 | 0.35 |
ENSDART00000046384
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr2_-_3611960 | 0.35 |
ENSDART00000184579
|
pter
|
phosphotriesterase related |
| chr10_-_26218354 | 0.34 |
ENSDART00000180764
|
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr2_-_40890264 | 0.34 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr4_+_33423119 | 0.33 |
ENSDART00000189413
|
znf1065
|
zinc finger protein 1065 |
| chr17_-_608857 | 0.33 |
ENSDART00000163431
|
KLHL28
|
kelch like family member 28 |
| chr11_-_36474306 | 0.33 |
ENSDART00000170678
ENSDART00000123591 |
usp48
|
ubiquitin specific peptidase 48 |
| chr5_-_26765188 | 0.32 |
ENSDART00000029450
|
rnf181
|
ring finger protein 181 |
| chr7_+_24390939 | 0.32 |
ENSDART00000087494
ENSDART00000125463 |
haus3
|
HAUS augmin-like complex, subunit 3 |
| chr14_+_23874062 | 0.32 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr2_-_8609653 | 0.32 |
ENSDART00000193354
ENSDART00000189489 ENSDART00000186144 |
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr8_+_7756893 | 0.31 |
ENSDART00000191894
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr8_+_12930216 | 0.31 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr5_+_51111766 | 0.31 |
ENSDART00000188552
|
pomt1
|
protein-O-mannosyltransferase 1 |
| chr19_-_11966015 | 0.31 |
ENSDART00000123409
|
si:ch1073-296d18.1
|
si:ch1073-296d18.1 |
| chr23_+_22291440 | 0.30 |
ENSDART00000185126
|
CR354612.1
|
|
| chr2_+_37424261 | 0.30 |
ENSDART00000132427
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr10_-_33265566 | 0.29 |
ENSDART00000063642
|
tbl2
|
transducin (beta)-like 2 |
| chr8_-_13823091 | 0.29 |
ENSDART00000177174
ENSDART00000137021 |
cabp4
|
calcium binding protein 4 |
| chr7_-_30560400 | 0.29 |
ENSDART00000142680
ENSDART00000142818 |
sltm
|
SAFB-like, transcription modulator |
| chr5_-_26764880 | 0.28 |
ENSDART00000140392
ENSDART00000134728 |
rnf181
|
ring finger protein 181 |
| chr8_-_43834442 | 0.28 |
ENSDART00000191927
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr7_-_59311165 | 0.28 |
ENSDART00000171105
|
m1ap
|
meiosis 1 associated protein |
| chr25_-_11016675 | 0.28 |
ENSDART00000099572
|
mespab
|
mesoderm posterior ab |
| chr1_+_47178529 | 0.28 |
ENSDART00000158432
ENSDART00000074450 ENSDART00000137448 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr16_+_13993285 | 0.27 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr2_+_55916911 | 0.27 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr20_+_25904199 | 0.27 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr19_-_26923957 | 0.26 |
ENSDART00000182390
|
skiv2l
|
SKI2 homolog, superkiller viralicidic activity 2-like |
| chr17_+_50074372 | 0.26 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr24_-_7587401 | 0.26 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr21_+_27513859 | 0.25 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr24_+_26017094 | 0.24 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr12_-_19862912 | 0.24 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr5_+_26765275 | 0.24 |
ENSDART00000144169
|
si:ch211-102c2.8
|
si:ch211-102c2.8 |
| chr24_-_11057305 | 0.24 |
ENSDART00000186494
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr22_+_1481307 | 0.24 |
ENSDART00000177429
|
si:ch211-255f4.2
|
si:ch211-255f4.2 |
| chr22_+_2598998 | 0.24 |
ENSDART00000176665
|
CU570894.1
|
|
| chrM_+_4993 | 0.23 |
ENSDART00000093600
|
mt-nd2
|
NADH dehydrogenase 2, mitochondrial |
| chr3_+_48234445 | 0.23 |
ENSDART00000161419
|
tbcd
|
tubulin folding cofactor D |
| chr19_+_4062101 | 0.22 |
ENSDART00000166773
|
btr25
|
bloodthirsty-related gene family, member 25 |
| chr20_-_5369105 | 0.22 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr4_+_45357558 | 0.21 |
ENSDART00000150769
|
si:ch211-162i8.5
|
si:ch211-162i8.5 |
| chr6_+_11250316 | 0.21 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr7_-_38664947 | 0.21 |
ENSDART00000100546
ENSDART00000112405 |
c6ast1
|
six-cysteine containing astacin protease 1 |
| chr11_-_8208464 | 0.21 |
ENSDART00000161283
|
pimr203
|
Pim proto-oncogene, serine/threonine kinase, related 203 |
| chr1_+_46598764 | 0.20 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr1_-_57129179 | 0.20 |
ENSDART00000157226
ENSDART00000152469 |
si:ch73-94k4.2
|
si:ch73-94k4.2 |
| chr22_-_36934040 | 0.20 |
ENSDART00000151666
|
pimr206
|
Pim proto-oncogene, serine/threonine kinase, related 206 |
| chr22_+_18389271 | 0.20 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr7_+_16352924 | 0.20 |
ENSDART00000158972
|
mpped2a
|
metallophosphoesterase domain containing 2a |
| chr2_-_37280028 | 0.20 |
ENSDART00000139459
|
nadkb
|
NAD kinase b |
| chr1_-_49950643 | 0.20 |
ENSDART00000138301
|
sgms2
|
sphingomyelin synthase 2 |
| chr4_-_65036768 | 0.19 |
ENSDART00000184455
|
si:ch211-283l16.1
|
si:ch211-283l16.1 |
| chr11_+_39928828 | 0.19 |
ENSDART00000137516
ENSDART00000134082 |
vamp3
|
vesicle-associated membrane protein 3 (cellubrevin) |
| chr3_-_15487111 | 0.19 |
ENSDART00000011320
|
nfatc2ip
|
nuclear factor of activated T cells 2 interacting protein |
| chr19_+_43885770 | 0.18 |
ENSDART00000135599
|
lypla2
|
lysophospholipase II |
| chr6_+_27991943 | 0.18 |
ENSDART00000143974
ENSDART00000141354 ENSDART00000088914 ENSDART00000139367 |
amotl2a
|
angiomotin like 2a |
| chr10_+_8550435 | 0.17 |
ENSDART00000185664
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr22_+_1330477 | 0.17 |
ENSDART00000157567
|
CU207221.2
|
|
| chr10_-_29831944 | 0.16 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr22_+_39058269 | 0.16 |
ENSDART00000113362
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr4_-_60049792 | 0.15 |
ENSDART00000158199
|
znf1033
|
zinc finger protein 1033 |
| chr1_+_58470287 | 0.14 |
ENSDART00000160245
ENSDART00000169945 |
si:ch73-236c18.7
|
si:ch73-236c18.7 |
| chr16_+_44906324 | 0.14 |
ENSDART00000074960
|
cd22
|
cd22 molecule |
| chr5_-_62317496 | 0.14 |
ENSDART00000180089
|
zgc:85789
|
zgc:85789 |
| chr22_-_10470663 | 0.14 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr22_+_36914636 | 0.14 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr19_-_6873107 | 0.14 |
ENSDART00000124440
|
CABZ01029822.1
|
|
| chr5_+_61843752 | 0.14 |
ENSDART00000130940
|
CR759879.1
|
Danio rerio interferon-induced protein with tetratricopeptide repeats 5 (LOC572297), mRNA. |
| chr11_-_29082429 | 0.14 |
ENSDART00000041443
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr14_+_36454087 | 0.14 |
ENSDART00000180247
|
TENM3
|
si:dkey-237h12.3 |
| chr22_-_5663354 | 0.14 |
ENSDART00000081774
|
ccdc51
|
coiled-coil domain containing 51 |
| chr3_-_111350 | 0.14 |
ENSDART00000171887
|
zgc:110249
|
zgc:110249 |
| chr2_+_269781 | 0.13 |
ENSDART00000156019
|
phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr13_+_28702104 | 0.13 |
ENSDART00000135481
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr7_+_26167420 | 0.12 |
ENSDART00000173941
|
si:ch211-196f2.6
|
si:ch211-196f2.6 |
| chr12_-_3453589 | 0.12 |
ENSDART00000175918
|
CABZ01063170.1
|
|
| chr5_+_43782267 | 0.12 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr10_-_24319526 | 0.12 |
ENSDART00000148480
|
inpp5kb
|
inositol polyphosphate-5-phosphatase Kb |
| chr14_+_16036139 | 0.12 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.4 | 1.1 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.3 | 1.0 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.3 | 0.8 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 0.7 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.2 | 0.6 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 0.9 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 0.7 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.2 | 0.6 | GO:0030237 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.2 | 0.6 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 0.6 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.2 | 1.0 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.2 | 0.5 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) mediolateral intercalation(GO:0060031) |
| 0.1 | 0.4 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 1.1 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.4 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.8 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.4 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 0.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.9 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.5 | GO:0060465 | pharynx development(GO:0060465) |
| 0.1 | 0.6 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.7 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.2 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.5 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.8 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.5 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.7 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 0.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.8 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.5 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.5 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.3 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0002370 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.4 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.5 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 1.1 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.0 | 0.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0035909 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0030823 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.4 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.5 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.1 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.0 | 0.1 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.8 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.3 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 1.0 | GO:0046834 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.6 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.8 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.6 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.9 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.6 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.2 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.6 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 0.8 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.4 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.3 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.9 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.4 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 1.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 1.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 1.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.8 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 5.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 0.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |