Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for nkx2.2b
Z-value: 0.58
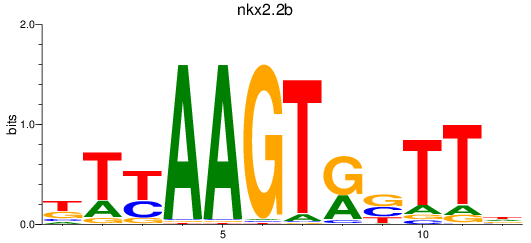
Transcription factors associated with nkx2.2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2b
|
ENSDARG00000101549 | NK2 homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2b | dr11_v1_chr20_+_48782068_48782068 | 0.27 | 4.9e-01 | Click! |
Activity profile of nkx2.2b motif
Sorted Z-values of nkx2.2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22158784 | 0.85 |
ENSDART00000167850
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr9_-_22158325 | 0.83 |
ENSDART00000114568
|
crygm2d16
|
crystallin, gamma M2d16 |
| chr8_+_26292560 | 0.79 |
ENSDART00000053463
|
mgll
|
monoglyceride lipase |
| chr9_-_18911608 | 0.78 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr14_-_25956804 | 0.74 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr23_+_6795531 | 0.73 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr4_+_842010 | 0.73 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr6_-_19341184 | 0.70 |
ENSDART00000168236
ENSDART00000167674 |
mif4gda
|
MIF4G domain containing a |
| chr19_-_24555935 | 0.67 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr5_-_17601759 | 0.67 |
ENSDART00000138387
|
si:ch211-130h14.6
|
si:ch211-130h14.6 |
| chr15_-_20468302 | 0.67 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr22_+_11775269 | 0.66 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr19_-_24555623 | 0.63 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr7_-_27038488 | 0.63 |
ENSDART00000052731
ENSDART00000191382 |
nucb2a
|
nucleobindin 2a |
| chr16_+_23913943 | 0.63 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr12_-_16595406 | 0.60 |
ENSDART00000166798
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr1_+_44442140 | 0.59 |
ENSDART00000190592
|
crybb1l2
|
crystallin, beta B1, like 2 |
| chr2_-_53525896 | 0.58 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr22_+_20720808 | 0.57 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr12_-_16595177 | 0.54 |
ENSDART00000133962
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr14_-_498979 | 0.54 |
ENSDART00000171976
|
spry1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr11_-_30636163 | 0.54 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr11_+_8660158 | 0.53 |
ENSDART00000169141
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr23_-_24394719 | 0.53 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr12_-_48671612 | 0.52 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr7_+_20109968 | 0.52 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr19_+_40861853 | 0.52 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr8_-_25074319 | 0.50 |
ENSDART00000134190
|
sort1b
|
sortilin 1b |
| chr25_-_29134000 | 0.49 |
ENSDART00000172027
ENSDART00000190447 |
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr6_-_41085443 | 0.49 |
ENSDART00000143944
ENSDART00000028217 |
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr3_-_15999501 | 0.49 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr7_-_38087865 | 0.48 |
ENSDART00000052366
|
cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr3_-_16000168 | 0.48 |
ENSDART00000160628
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr6_-_13408680 | 0.46 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr21_-_37733571 | 0.44 |
ENSDART00000176214
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr14_-_42997145 | 0.43 |
ENSDART00000172801
|
pcdh10b
|
protocadherin 10b |
| chr6_-_41085692 | 0.43 |
ENSDART00000181463
|
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr13_+_44857087 | 0.43 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr14_+_21861099 | 0.43 |
ENSDART00000032141
|
spon2a
|
spondin 2a, extracellular matrix protein |
| chr20_+_33904258 | 0.43 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr12_-_26407092 | 0.42 |
ENSDART00000178687
|
myoz1b
|
myozenin 1b |
| chr3_+_24134418 | 0.42 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr21_+_26389391 | 0.41 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr19_+_38422059 | 0.41 |
ENSDART00000035093
|
col9a2
|
procollagen, type IX, alpha 2 |
| chr12_+_6002715 | 0.41 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr1_+_15137901 | 0.40 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr2_-_32352946 | 0.40 |
ENSDART00000144870
ENSDART00000077151 |
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr23_+_6795709 | 0.39 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr15_+_13984879 | 0.38 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr4_+_2092683 | 0.38 |
ENSDART00000067423
|
frs2a
|
fibroblast growth factor receptor substrate 2a |
| chr23_-_4925641 | 0.38 |
ENSDART00000140861
ENSDART00000060718 |
taz
|
tafazzin |
| chr4_+_26449406 | 0.38 |
ENSDART00000183291
|
CU137645.3
|
|
| chr23_+_19813677 | 0.38 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr2_+_26498446 | 0.37 |
ENSDART00000078412
|
rps8a
|
ribosomal protein S8a |
| chr10_-_7885654 | 0.37 |
ENSDART00000185768
|
BX294661.2
|
|
| chr7_-_16562200 | 0.37 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr16_+_4658250 | 0.36 |
ENSDART00000006212
|
LIN28A
|
si:ch1073-284b18.2 |
| chr7_-_56793739 | 0.36 |
ENSDART00000082842
|
si:ch211-146m13.3
|
si:ch211-146m13.3 |
| chr6_-_7439490 | 0.36 |
ENSDART00000188825
|
fkbp11
|
FK506 binding protein 11 |
| chr5_-_47727819 | 0.36 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr12_-_8511185 | 0.36 |
ENSDART00000179910
|
CR932364.1
|
|
| chr19_+_47135677 | 0.36 |
ENSDART00000139692
|
si:ch211-229b6.1
|
si:ch211-229b6.1 |
| chr16_+_8716800 | 0.36 |
ENSDART00000124693
ENSDART00000181961 |
cabz01093075.1
|
cabz01093075.1 |
| chr12_+_9880493 | 0.36 |
ENSDART00000055019
|
ndufa4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 |
| chr1_+_36651059 | 0.36 |
ENSDART00000187475
|
ednraa
|
endothelin receptor type Aa |
| chr21_-_40835069 | 0.35 |
ENSDART00000004686
|
limk1b
|
LIM domain kinase 1b |
| chr1_+_52560549 | 0.35 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr23_+_25354856 | 0.35 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr20_+_492529 | 0.35 |
ENSDART00000141709
|
FO904980.1
|
|
| chr5_-_58939460 | 0.35 |
ENSDART00000122413
|
mcama
|
melanoma cell adhesion molecule a |
| chr7_-_69636502 | 0.34 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr9_-_23156908 | 0.33 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr5_-_60159116 | 0.33 |
ENSDART00000147675
|
si:dkey-280e8.1
|
si:dkey-280e8.1 |
| chr25_-_16755340 | 0.33 |
ENSDART00000124729
ENSDART00000110859 |
ribc2
|
RIB43A domain with coiled-coils 2 |
| chr3_-_27647845 | 0.33 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr16_-_51288178 | 0.33 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr18_+_37272568 | 0.32 |
ENSDART00000132749
|
tmem123
|
transmembrane protein 123 |
| chr18_-_40481028 | 0.32 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr14_-_10617923 | 0.32 |
ENSDART00000133723
ENSDART00000131939 ENSDART00000136649 |
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr1_+_45351890 | 0.32 |
ENSDART00000145486
|
si:ch211-243a20.3
|
si:ch211-243a20.3 |
| chr3_-_26122353 | 0.31 |
ENSDART00000111042
ENSDART00000161427 |
hsp70.2
|
heat shock cognate 70-kd protein, tandem duplicate 2 |
| chr4_+_6032640 | 0.31 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr4_+_33225161 | 0.31 |
ENSDART00000150851
|
si:dkey-247i3.5
|
si:dkey-247i3.5 |
| chr13_-_43238578 | 0.30 |
ENSDART00000161017
|
eif3s6ip
|
eukaryotic translation initiation factor 3, subunit 6 interacting protein |
| chr6_+_49255706 | 0.30 |
ENSDART00000156866
|
si:dkey-183k8.2
|
si:dkey-183k8.2 |
| chr3_+_28581397 | 0.29 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr16_+_32995882 | 0.29 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr4_+_58576146 | 0.29 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr14_+_18781082 | 0.28 |
ENSDART00000108793
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr12_+_30233977 | 0.28 |
ENSDART00000188135
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr19_-_22328154 | 0.28 |
ENSDART00000090464
|
si:ch73-196l6.5
|
si:ch73-196l6.5 |
| chr16_-_11798994 | 0.27 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr1_+_12767318 | 0.27 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr12_+_30234209 | 0.27 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr16_+_16933002 | 0.27 |
ENSDART00000173163
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr15_-_1198886 | 0.27 |
ENSDART00000063285
|
lxn
|
latexin |
| chr21_-_42055872 | 0.27 |
ENSDART00000144767
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr23_-_17470146 | 0.27 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr9_+_21722733 | 0.26 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr21_+_9628854 | 0.26 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr10_+_44984946 | 0.26 |
ENSDART00000182520
|
h2afvb
|
H2A histone family, member Vb |
| chr21_+_33454147 | 0.26 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr4_+_28356606 | 0.25 |
ENSDART00000192995
|
si:ch73-263o4.4
|
si:ch73-263o4.4 |
| chr4_+_62184754 | 0.25 |
ENSDART00000168844
|
si:dkeyp-35e5.9
|
si:dkeyp-35e5.9 |
| chr19_-_35010226 | 0.25 |
ENSDART00000051750
ENSDART00000155525 |
si:rp71-45k5.4
|
si:rp71-45k5.4 |
| chr25_+_10485103 | 0.24 |
ENSDART00000104576
|
psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr4_-_25836684 | 0.24 |
ENSDART00000142491
|
ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr7_+_34231782 | 0.24 |
ENSDART00000173547
|
lctla
|
lactase-like a |
| chr3_+_60716904 | 0.24 |
ENSDART00000168280
|
foxj1a
|
forkhead box J1a |
| chr23_+_36653376 | 0.24 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr16_+_23087326 | 0.24 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr4_-_22311610 | 0.24 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr9_+_25776971 | 0.24 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr7_+_29992889 | 0.24 |
ENSDART00000055936
|
isl2b
|
ISL LIM homeobox 2b |
| chr20_-_30920356 | 0.24 |
ENSDART00000022951
|
kif25
|
kinesin family member 25 |
| chr14_-_10371638 | 0.24 |
ENSDART00000003879
|
fam199x
|
family with sequence similarity 199, X-linked |
| chr11_+_24900123 | 0.24 |
ENSDART00000044987
ENSDART00000148023 |
timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr19_+_40856807 | 0.23 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr12_-_35054354 | 0.23 |
ENSDART00000075351
|
zgc:112285
|
zgc:112285 |
| chr4_-_9891874 | 0.23 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr4_+_58016732 | 0.23 |
ENSDART00000165777
|
CT027609.1
|
|
| chr2_-_21618629 | 0.23 |
ENSDART00000027587
|
ralab
|
v-ral simian leukemia viral oncogene homolog Ab (ras related) |
| chr2_-_3038904 | 0.23 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr10_-_27197044 | 0.23 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr16_+_20915319 | 0.22 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr19_-_19339285 | 0.22 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr8_-_14484599 | 0.22 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr25_-_22363253 | 0.22 |
ENSDART00000073573
ENSDART00000162103 |
ccdc33
|
coiled-coil domain containing 33 |
| chr5_-_28052883 | 0.22 |
ENSDART00000188791
|
zgc:113436
|
zgc:113436 |
| chr20_-_4766645 | 0.22 |
ENSDART00000147071
ENSDART00000152398 |
ak7a
|
adenylate kinase 7a |
| chr11_+_18873113 | 0.22 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr21_+_30549512 | 0.22 |
ENSDART00000132831
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr10_-_44924289 | 0.22 |
ENSDART00000171267
|
tuba7l
|
tubulin, alpha 7 like |
| chr22_-_20720427 | 0.21 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr8_+_14058646 | 0.21 |
ENSDART00000080852
|
ugt5e1
|
UDP glucuronosyltransferase 5 family, polypeptide E1 |
| chr4_-_5333971 | 0.21 |
ENSDART00000067375
|
osgep
|
O-sialoglycoprotein endopeptidase |
| chr19_-_48330287 | 0.21 |
ENSDART00000162244
|
si:ch73-359m17.7
|
si:ch73-359m17.7 |
| chr12_+_30168342 | 0.21 |
ENSDART00000142756
|
ablim1b
|
actin binding LIM protein 1b |
| chr9_+_23900703 | 0.21 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr2_-_37956768 | 0.21 |
ENSDART00000034595
|
cbln10
|
cerebellin 10 |
| chr2_-_7696287 | 0.21 |
ENSDART00000190769
|
CABZ01055306.1
|
|
| chr1_+_23784905 | 0.21 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr9_+_55056310 | 0.21 |
ENSDART00000047080
|
gpr143
|
G protein-coupled receptor 143 |
| chr8_+_22582146 | 0.20 |
ENSDART00000157655
ENSDART00000189892 |
CT583651.2
|
|
| chr10_-_20706109 | 0.20 |
ENSDART00000124751
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr12_+_36109507 | 0.20 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr4_-_25215968 | 0.20 |
ENSDART00000066932
ENSDART00000066933 |
itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr4_-_72609735 | 0.20 |
ENSDART00000174299
ENSDART00000159227 |
si:cabz01054394.6
|
si:cabz01054394.6 |
| chr1_+_33328857 | 0.20 |
ENSDART00000137151
|
mxra5a
|
matrix-remodelling associated 5a |
| chr22_-_4892937 | 0.20 |
ENSDART00000147002
|
si:ch73-256j6.4
|
si:ch73-256j6.4 |
| chr10_+_9372702 | 0.19 |
ENSDART00000021100
|
lhx6
|
LIM homeobox 6 |
| chr8_+_25302172 | 0.19 |
ENSDART00000046182
ENSDART00000145316 |
gstm.3
|
glutathione S-transferase mu tandem duplicate 3 |
| chr5_-_9073433 | 0.19 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr7_-_35083184 | 0.19 |
ENSDART00000100253
ENSDART00000135250 ENSDART00000173511 |
agrp
|
agouti related neuropeptide |
| chr6_-_13187168 | 0.19 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr12_-_30548244 | 0.19 |
ENSDART00000193616
|
zgc:158404
|
zgc:158404 |
| chr15_+_5299404 | 0.19 |
ENSDART00000155410
|
or122-2
|
odorant receptor, family E, subfamily 122, member 2 |
| chr4_+_4267451 | 0.19 |
ENSDART00000192069
|
ano2
|
anoctamin 2 |
| chr8_-_9118958 | 0.19 |
ENSDART00000037922
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr2_+_1487118 | 0.18 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr24_+_5935377 | 0.18 |
ENSDART00000191989
ENSDART00000185932 ENSDART00000131768 |
abi1a
|
abl-interactor 1a |
| chr6_-_13200585 | 0.18 |
ENSDART00000185321
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr21_+_25793970 | 0.18 |
ENSDART00000101217
|
CLDN4 (1 of many)
|
zgc:136892 |
| chr14_+_17376940 | 0.18 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr4_+_65614183 | 0.18 |
ENSDART00000180387
|
si:dkey-205i10.1
|
si:dkey-205i10.1 |
| chr19_-_22478888 | 0.18 |
ENSDART00000090679
|
pleca
|
plectin a |
| chr4_-_17838179 | 0.17 |
ENSDART00000146931
|
avpr2l
|
arginine vasopressin receptor 2, like |
| chr4_+_31405646 | 0.17 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr7_+_57108823 | 0.17 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr21_+_44112914 | 0.17 |
ENSDART00000062836
|
fgf1b
|
fibroblast growth factor 1b |
| chr15_+_31526225 | 0.17 |
ENSDART00000154456
|
wdr95
|
WD40 repeat domain 95 |
| chr3_-_7524363 | 0.17 |
ENSDART00000162970
|
znf1001
|
zinc finger protein 1001 |
| chr11_+_30314885 | 0.17 |
ENSDART00000187418
ENSDART00000123244 |
ugt1b2
|
UDP glucuronosyltransferase 1 family, polypeptide B2 |
| chr18_-_6766354 | 0.17 |
ENSDART00000132611
|
adm2b
|
adrenomedullin 2b |
| chr10_+_21722892 | 0.17 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr2_-_1514001 | 0.17 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr16_-_41465542 | 0.17 |
ENSDART00000169116
ENSDART00000187446 |
cpne4a
|
copine IVa |
| chr6_-_57938043 | 0.17 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr2_-_59376735 | 0.17 |
ENSDART00000193624
|
ftr38
|
finTRIM family, member 38 |
| chr12_-_25612170 | 0.17 |
ENSDART00000077155
|
six2b
|
SIX homeobox 2b |
| chr15_+_24691088 | 0.16 |
ENSDART00000110618
|
LRRC75A
|
si:dkey-151p21.7 |
| chr5_-_67499279 | 0.16 |
ENSDART00000128050
|
si:dkey-251i10.3
|
si:dkey-251i10.3 |
| chr14_-_858985 | 0.16 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr22_+_30839702 | 0.16 |
ENSDART00000139849
ENSDART00000135775 |
si:dkey-49n23.3
|
si:dkey-49n23.3 |
| chr19_-_677713 | 0.16 |
ENSDART00000025146
|
slc6a19a.1
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 1 |
| chr4_+_69823638 | 0.16 |
ENSDART00000165786
|
znf1087
|
zinc finger protein 1087 |
| chr2_-_51757328 | 0.16 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr4_+_43679421 | 0.16 |
ENSDART00000132945
|
zgc:173705
|
zgc:173705 |
| chr1_-_46832880 | 0.16 |
ENSDART00000142406
|
si:ch73-160h15.3
|
si:ch73-160h15.3 |
| chr5_-_37900350 | 0.16 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr24_-_36680261 | 0.16 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr2_-_32386942 | 0.16 |
ENSDART00000131408
|
ubtfl
|
upstream binding transcription factor, like |
| chr20_-_32188897 | 0.16 |
ENSDART00000133887
|
si:ch211-51a19.5
|
si:ch211-51a19.5 |
| chr22_+_1317353 | 0.15 |
ENSDART00000187250
|
si:ch73-138e16.6
|
si:ch73-138e16.6 |
| chr23_+_1078072 | 0.15 |
ENSDART00000159263
ENSDART00000053527 |
slc34a2b
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 2b |
| chr5_+_26795773 | 0.15 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr23_-_19827411 | 0.15 |
ENSDART00000187964
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr18_-_14691727 | 0.15 |
ENSDART00000010129
|
pdf
|
peptide deformylase, mitochondrial |
| chr4_+_76441823 | 0.15 |
ENSDART00000163932
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr19_-_29832876 | 0.15 |
ENSDART00000005119
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.2 | 0.5 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.1 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.7 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.4 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.4 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.2 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.6 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 1.0 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.4 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.2 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.2 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.4 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.4 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.2 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.7 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.5 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.4 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.5 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.2 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0015893 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0060092 | inhibitory postsynaptic potential(GO:0060080) regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 2.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.2 | GO:0030323 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.0 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.1 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.1 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.3 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.4 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.2 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 1.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 2.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.6 | GO:0019956 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |